ZFLNCG04876
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR592703 | pineal gland | normal | 606.12 |
| SRR592702 | pineal gland | normal | 448.78 |
| SRR592699 | pineal gland | normal | 194.76 |
| ERR023144 | brain | normal | 174.85 |
| SRR592701 | pineal gland | normal | 172.13 |
| SRR519727 | 6 dpf | FETOH treatment | 150.27 |
| SRR519747 | 6 dpf | VD3 treatment | 131.30 |
| SRR519752 | 7 dpf | VD3 treatment | 129.97 |
| SRR1048061 | pineal gland | dark | 126.39 |
| SRR592698 | pineal gland | normal | 99.64 |
Express in tissues
Correlated coding gene
Download
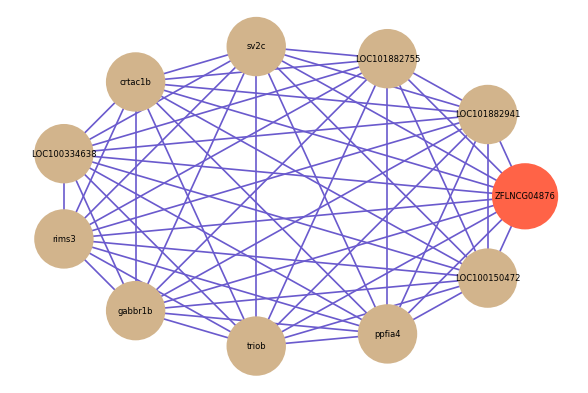
| gene | correlation coefficent |
|---|---|
| LOC101882941 | 0.78 |
| LOC101882755 | 0.76 |
| sv2c | 0.74 |
| crtac1b | 0.73 |
| LOC100334638 | 0.73 |
| rims3 | 0.73 |
| gabbr1b | 0.73 |
| triob | 0.73 |
| ppfia4 | 0.73 |
| LOC100150472 | 0.73 |
Gene Ontology
Download
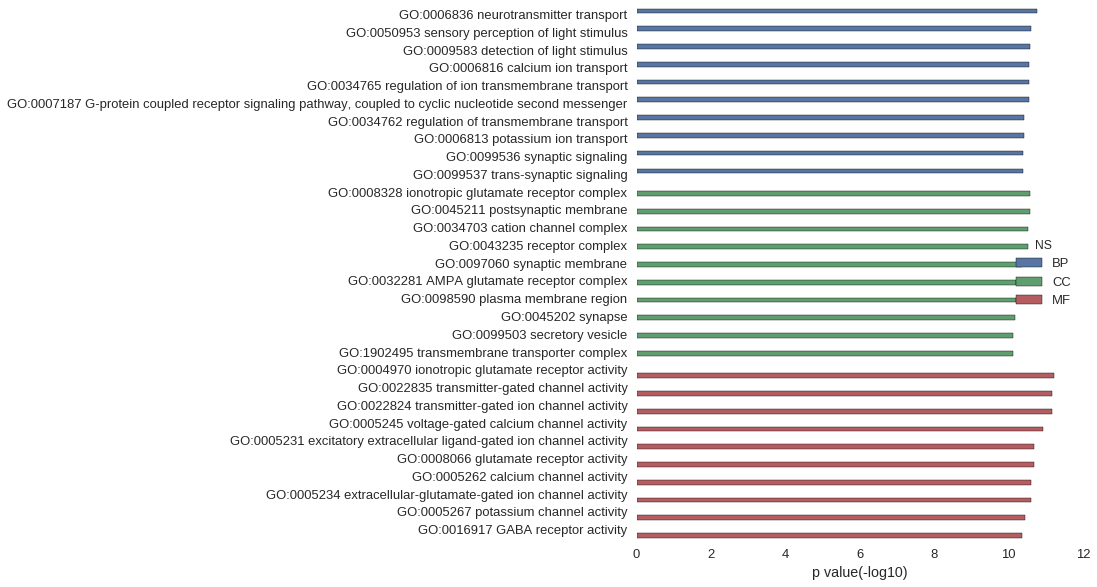
| GO | P value |
|---|---|
| GO:0006836 | 1.68e-11 |
| GO:0050953 | 2.45e-11 |
| GO:0009583 | 2.70e-11 |
| GO:0006816 | 2.76e-11 |
| GO:0034765 | 2.77e-11 |
| GO:0007187 | 2.87e-11 |
| GO:0034762 | 3.96e-11 |
| GO:0006813 | 3.96e-11 |
| GO:0099536 | 4.11e-11 |
| GO:0099537 | 4.11e-11 |
| GO:0008328 | 2.63e-11 |
| GO:0045211 | 2.70e-11 |
| GO:0034703 | 2.91e-11 |
| GO:0043235 | 2.93e-11 |
| GO:0097060 | 4.39e-11 |
| GO:0032281 | 6.29e-11 |
| GO:0098590 | 6.40e-11 |
| GO:0045202 | 6.61e-11 |
| GO:0099503 | 7.45e-11 |
| GO:1902495 | 7.81e-11 |
| GO:0004970 | 5.89e-12 |
| GO:0022835 | 6.93e-12 |
| GO:0022824 | 6.93e-12 |
| GO:0005245 | 1.16e-11 |
| GO:0005231 | 2.10e-11 |
| GO:0008066 | 2.12e-11 |
| GO:0005262 | 2.45e-11 |
| GO:0005234 | 2.49e-11 |
| GO:0005267 | 3.50e-11 |
| GO:0016917 | 4.22e-11 |
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG04876
ATAGATAGAGGTAGATAGATAGATAGATATTTAATCAATAAATGAAACGTGACTCACCTAGAGTTGCAGT
CTCCAGCAGACTGGCCAATAACAGCACTTGTAAAATCCCCATGGCCCCACCCCTCACTCTGGATTAGCTG
GTTTGGGCTGAGCTCCTCCCCTTAGAGTGGGTTGATTGGCCGTATATAAAGCGACTGCACTCAGATCAGA
GTTCCTGTTTGTCTGAAAACAGCAAAGCCACCTATGAACACAGAGAGAGA
ATAGATAGAGGTAGATAGATAGATAGATATTTAATCAATAAATGAAACGTGACTCACCTAGAGTTGCAGT
CTCCAGCAGACTGGCCAATAACAGCACTTGTAAAATCCCCATGGCCCCACCCCTCACTCTGGATTAGCTG
GTTTGGGCTGAGCTCCTCCCCTTAGAGTGGGTTGATTGGCCGTATATAAAGCGACTGCACTCAGATCAGA
GTTCCTGTTTGTCTGAAAACAGCAAAGCCACCTATGAACACAGAGAGAGA