ZFLNCG05477
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR023144 | brain | normal | 41.22 |
| SRR527833 | 5 dpf | normal | 12.47 |
| SRR1648855 | brain | normal | 11.89 |
| SRR726541 | 5 dpf | infection with Mycobacterium marinum | 10.51 |
| SRR726540 | 5 dpf | normal | 9.70 |
| SRR1291417 | 5 dpi | injected marinum | 9.61 |
| SRR519732 | 7 dpf | FETOH treatment | 9.54 |
| SRR519752 | 7 dpf | VD3 treatment | 9.40 |
| SRR519727 | 6 dpf | FETOH treatment | 8.66 |
| SRR891511 | brain | normal | 8.53 |
Express in tissues
Correlated coding gene
Download
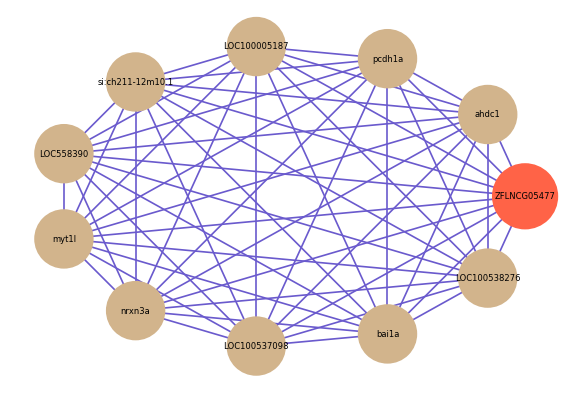
| gene | correlation coefficent |
|---|---|
| ahdc1 | 0.84 |
| pcdh1a | 0.83 |
| LOC100005187 | 0.83 |
| si:ch211-12m10.1 | 0.83 |
| LOC558390 | 0.82 |
| myt1l | 0.82 |
| nrxn3a | 0.82 |
| LOC100537098 | 0.82 |
| bai1a | 0.82 |
| LOC100538276 | 0.82 |
Gene Ontology
Download
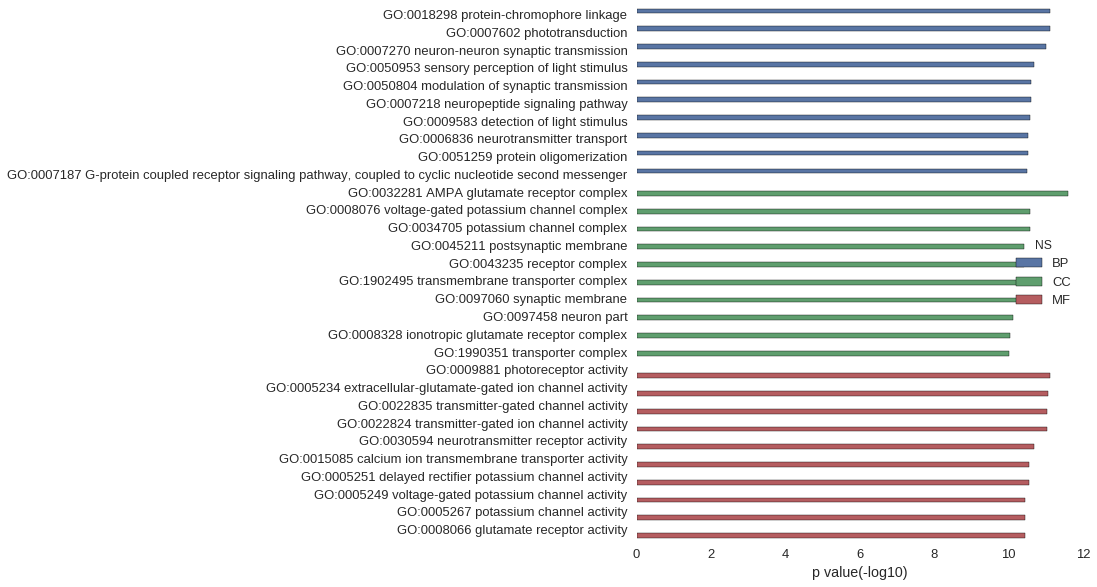
| GO | P value |
|---|---|
| GO:0018298 | 7.63e-12 |
| GO:0007602 | 7.63e-12 |
| GO:0007270 | 9.61e-12 |
| GO:0050953 | 2.03e-11 |
| GO:0050804 | 2.53e-11 |
| GO:0007218 | 2.55e-11 |
| GO:0009583 | 2.61e-11 |
| GO:0006836 | 3.00e-11 |
| GO:0051259 | 3.02e-11 |
| GO:0007187 | 3.27e-11 |
| GO:0032281 | 2.55e-12 |
| GO:0008076 | 2.61e-11 |
| GO:0034705 | 2.61e-11 |
| GO:0045211 | 3.84e-11 |
| GO:0043235 | 3.90e-11 |
| GO:1902495 | 5.26e-11 |
| GO:0097060 | 5.48e-11 |
| GO:0097458 | 7.73e-11 |
| GO:0008328 | 9.01e-11 |
| GO:1990351 | 9.48e-11 |
| GO:0009881 | 7.63e-12 |
| GO:0005234 | 8.75e-12 |
| GO:0022835 | 9.03e-12 |
| GO:0022824 | 9.03e-12 |
| GO:0030594 | 2.10e-11 |
| GO:0015085 | 2.75e-11 |
| GO:0005251 | 2.78e-11 |
| GO:0005249 | 3.50e-11 |
| GO:0005267 | 3.53e-11 |
| GO:0008066 | 3.64e-11 |
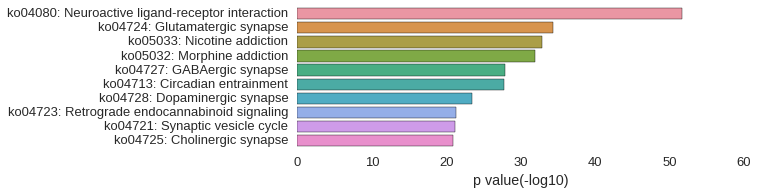
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG05477
TCTTCTGGCTCTGCTGTTGGGCTGTGAACACACGTACACGATTCCCAAATCATCTGAGGACAACAATGAT
GCAGGCGTTACTTTATGATGAGCTTTATTCATAATGAAGTGTTCCTGATTAAGACTTTAGGTGTGTAACA
GTGCGATTATTCGTGATGCTTACAGTCTGGCTGTCAGAGCGAGCGGTGTTGTCTTCTGTGTATTATGACC
CGGACCTTGACTTGGTGGAGTCCTGTGATTCCTGTTCTGGTTGGCTGAAAATACAAGCATGCATGAGTGC
ATATATGTACACACACACACACACACATAAGCATACATACACATACACAAATGCCCACATGCATGCAGGC
ACATGCCAACAAAAACACATG
TCTTCTGGCTCTGCTGTTGGGCTGTGAACACACGTACACGATTCCCAAATCATCTGAGGACAACAATGAT
GCAGGCGTTACTTTATGATGAGCTTTATTCATAATGAAGTGTTCCTGATTAAGACTTTAGGTGTGTAACA
GTGCGATTATTCGTGATGCTTACAGTCTGGCTGTCAGAGCGAGCGGTGTTGTCTTCTGTGTATTATGACC
CGGACCTTGACTTGGTGGAGTCCTGTGATTCCTGTTCTGGTTGGCTGAAAATACAAGCATGCATGAGTGC
ATATATGTACACACACACACACACACATAAGCATACATACACATACACAAATGCCCACATGCATGCAGGC
ACATGCCAACAAAAACACATG