ZFLNCG05551
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR065196 | 3 dpf | normal | 21.96 |
| SRR519717 | 2 dpf | FETOH treatment | 17.20 |
| SRR519737 | 2 dpf | VD3 treatment | 16.58 |
| SRR065197 | 3 dpf | U1C knockout | 14.07 |
| SRR610742 | 3 dpf | express NICD | 10.10 |
| SRR610741 | 3 dpf | express Ras | 9.59 |
| SRR831692 | embryo | normal | 9.10 |
| SRR1523214 | embryo | glut12 morpholino | 9.08 |
| SRR519742 | 4 dpf | VD3 treatment | 9.00 |
| SRR372802 | 5 dpf | normal | 8.93 |
Express in tissues
Correlated coding gene
Download
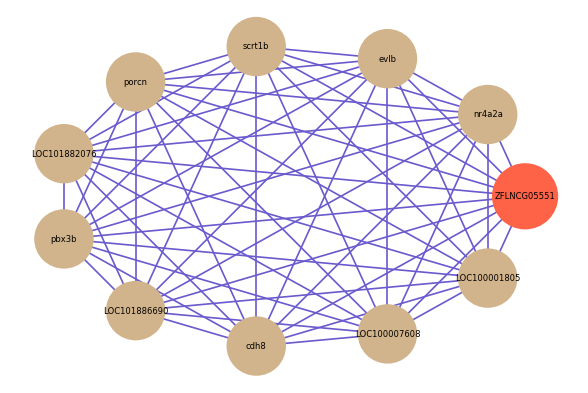
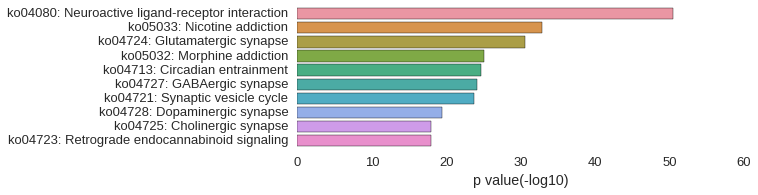
| gene | correlation coefficent |
|---|---|
| nr4a2a | 0.90 |
| evlb | 0.85 |
| scrt1b | 0.84 |
| porcn | 0.84 |
| LOC101882076 | 0.83 |
| pbx3b | 0.83 |
| LOC101886690 | 0.83 |
| cdh8 | 0.83 |
| LOC100007608 | 0.83 |
| LOC100001805 | 0.83 |
Gene Ontology
Download
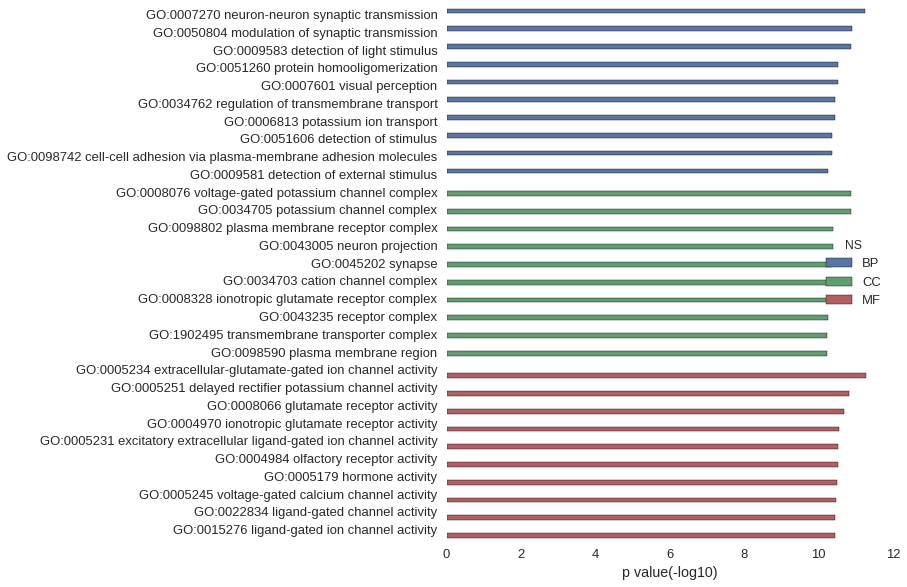
| GO | P value |
|---|---|
| GO:0007270 | 5.61e-12 |
| GO:0050804 | 1.26e-11 |
| GO:0009583 | 1.36e-11 |
| GO:0051260 | 2.94e-11 |
| GO:0007601 | 3.03e-11 |
| GO:0034762 | 3.55e-11 |
| GO:0006813 | 3.55e-11 |
| GO:0051606 | 4.24e-11 |
| GO:0098742 | 4.46e-11 |
| GO:0009581 | 5.41e-11 |
| GO:0008076 | 1.36e-11 |
| GO:0034705 | 1.36e-11 |
| GO:0098802 | 4.04e-11 |
| GO:0043005 | 4.04e-11 |
| GO:0045202 | 4.46e-11 |
| GO:0034703 | 4.47e-11 |
| GO:0008328 | 5.06e-11 |
| GO:0043235 | 5.46e-11 |
| GO:1902495 | 6.07e-11 |
| GO:0098590 | 6.09e-11 |
| GO:0005234 | 5.24e-12 |
| GO:0005251 | 1.55e-11 |
| GO:0008066 | 2.09e-11 |
| GO:0004970 | 2.88e-11 |
| GO:0005231 | 2.95e-11 |
| GO:0004984 | 2.95e-11 |
| GO:0005179 | 3.19e-11 |
| GO:0005245 | 3.36e-11 |
| GO:0022834 | 3.59e-11 |
| GO:0015276 | 3.59e-11 |
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG05551
TGATTTCGGTATTGGTCAAGTCCATGCTGAACTTCACAAACTCTGGCGTGAGGAAATCGCAGCTGTATTC
TCCGCTTGTGTGGTAACTGTAGCTCTGAGACGCAGGACTAGCGCCTTGCGGAGATGAGCCATACTGAGCC
TGGACGCAGGGCATGGCTGCAATTAAAACAAACAAAAAAATAAATAAATAAACAGAAAGACAATCATAAA
AAACAATAAATAAAATAAACACGCATTCCCCTAATAACACGTTTTCAGCTTGCATGCAACCTTAGCAAAT
TGATTTCAACGTAAGTACCTTCAGTCGTCGGACTGTTCCTCAGCAGATCTCGGCGTAGGTGATATTATCA
GGTGAAATTTAATACAGGGTTCACGACAAACGAATCAGTTCTGGATTCTGCCTCTGATAAAACACATAAC
TTTCACTTGATTTTGCATCACATTTGATTACTTCCTTAAGCCTACATAATTATGCAATACTCCAAAGCTC
ACTTAAACTTTGTATGCATATTAAAGCGAAGTTTTGCGCGCAGCGTTGACATGTTTTATTCTATAGTTTA
ATGCTTTTACATTCTTAGAATAAATAAATACATCTAAATAAAATAGTTTAAAATTTATAGTTTATAATAA
AATAGTTTCAGACAATAAATAAATAAAAGCTTAAAAGTTCACATTGCAGTCTTAGTATATATATATATAT
ATATATATATATATATATATATATATATATATATATATATATATATATATATATATACACACACACATAC
ACACACACACACACACACACACATACACACAACAAACGGACAAACGCAAATCTGTTTAAAACAAACATGC
AATGGGACAATTGCACTATACACAGAATAAAAAGACACTAACATACCTTAATAGAACATCATCAGTAAAA
CAATATCGTATCCATATGCTTCTGTATGGCGGTGATGGAGAATATGTGTATAAATGTCCAGTATAGCGTT
GTCCGCCGGACCCGGAGCTCAGCACTGGCATGTA
TGATTTCGGTATTGGTCAAGTCCATGCTGAACTTCACAAACTCTGGCGTGAGGAAATCGCAGCTGTATTC
TCCGCTTGTGTGGTAACTGTAGCTCTGAGACGCAGGACTAGCGCCTTGCGGAGATGAGCCATACTGAGCC
TGGACGCAGGGCATGGCTGCAATTAAAACAAACAAAAAAATAAATAAATAAACAGAAAGACAATCATAAA
AAACAATAAATAAAATAAACACGCATTCCCCTAATAACACGTTTTCAGCTTGCATGCAACCTTAGCAAAT
TGATTTCAACGTAAGTACCTTCAGTCGTCGGACTGTTCCTCAGCAGATCTCGGCGTAGGTGATATTATCA
GGTGAAATTTAATACAGGGTTCACGACAAACGAATCAGTTCTGGATTCTGCCTCTGATAAAACACATAAC
TTTCACTTGATTTTGCATCACATTTGATTACTTCCTTAAGCCTACATAATTATGCAATACTCCAAAGCTC
ACTTAAACTTTGTATGCATATTAAAGCGAAGTTTTGCGCGCAGCGTTGACATGTTTTATTCTATAGTTTA
ATGCTTTTACATTCTTAGAATAAATAAATACATCTAAATAAAATAGTTTAAAATTTATAGTTTATAATAA
AATAGTTTCAGACAATAAATAAATAAAAGCTTAAAAGTTCACATTGCAGTCTTAGTATATATATATATAT
ATATATATATATATATATATATATATATATATATATATATATATATATATATATATACACACACACATAC
ACACACACACACACACACACACATACACACAACAAACGGACAAACGCAAATCTGTTTAAAACAAACATGC
AATGGGACAATTGCACTATACACAGAATAAAAAGACACTAACATACCTTAATAGAACATCATCAGTAAAA
CAATATCGTATCCATATGCTTCTGTATGGCGGTGATGGAGAATATGTGTATAAATGTCCAGTATAGCGTT
GTCCGCCGGACCCGGAGCTCAGCACTGGCATGTA