ZFLNCG05646
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR592703 | pineal gland | normal | 132.06 |
| SRR592702 | pineal gland | normal | 86.16 |
| ERR023145 | heart | normal | 78.77 |
| SRR592700 | pineal gland | normal | 77.10 |
| SRR592701 | pineal gland | normal | 59.01 |
| SRR1647684 | spleen | SVCV treatment | 47.88 |
| ERR023146 | head kidney | normal | 45.09 |
| ERR023143 | swim bladder | normal | 38.85 |
| SRR592699 | pineal gland | normal | 35.52 |
| SRR516123 | skin | male and 3.5 year | 30.29 |
Express in tissues
Correlated coding gene
Download
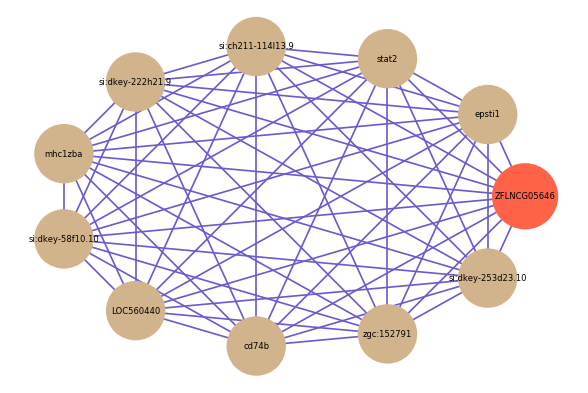
| gene | correlation coefficent |
|---|---|
| epsti1 | 0.85 |
| stat2 | 0.85 |
| si:ch211-114l13.9 | 0.84 |
| si:dkey-222h21.9 | 0.83 |
| mhc1zba | 0.83 |
| si:dkey-58f10.10 | 0.83 |
| LOC560440 | 0.83 |
| cd74b | 0.82 |
| zgc:152791 | 0.82 |
| si:dkey-253d23.10 | 0.82 |
Gene Ontology
Download
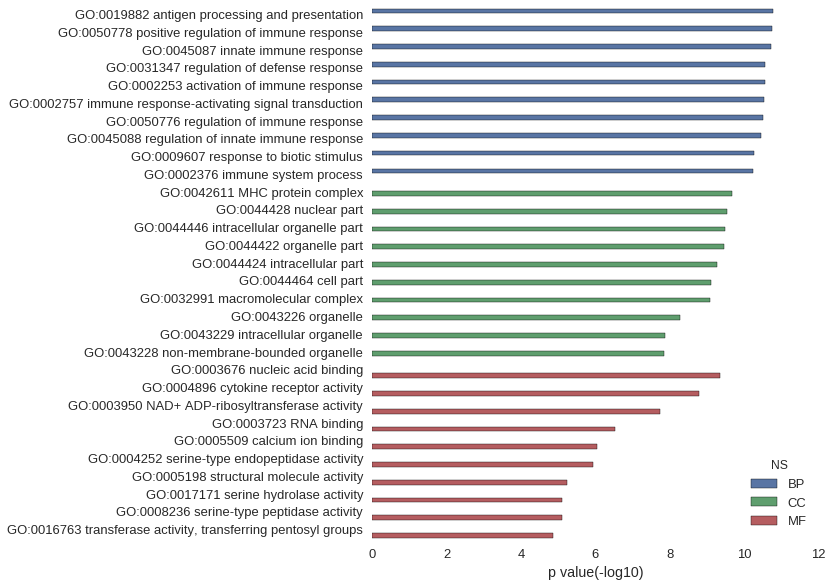
| GO | P value |
|---|---|
| GO:0019882 | 1.69e-11 |
| GO:0050778 | 1.81e-11 |
| GO:0045087 | 1.94e-11 |
| GO:0031347 | 2.71e-11 |
| GO:0002253 | 2.71e-11 |
| GO:0002757 | 2.95e-11 |
| GO:0050776 | 3.07e-11 |
| GO:0045088 | 3.55e-11 |
| GO:0009607 | 5.45e-11 |
| GO:0002376 | 5.67e-11 |
| GO:0042611 | 2.06e-10 |
| GO:0044428 | 2.81e-10 |
| GO:0044446 | 3.34e-10 |
| GO:0044422 | 3.47e-10 |
| GO:0044424 | 5.40e-10 |
| GO:0044464 | 7.62e-10 |
| GO:0032991 | 8.15e-10 |
| GO:0043226 | 5.31e-09 |
| GO:0043229 | 1.35e-08 |
| GO:0043228 | 1.46e-08 |
| GO:0003676 | 4.44e-10 |
| GO:0004896 | 1.60e-09 |
| GO:0003950 | 1.82e-08 |
| GO:0003723 | 3.02e-07 |
| GO:0005509 | 8.78e-07 |
| GO:0004252 | 1.16e-06 |
| GO:0005198 | 5.63e-06 |
| GO:0017171 | 7.84e-06 |
| GO:0008236 | 7.84e-06 |
| GO:0016763 | 1.40e-05 |
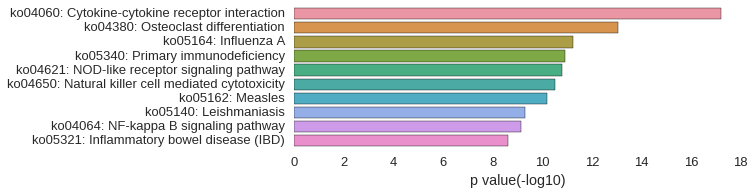
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG05646
CCGGAACACAACTGTTGCACTCTATTTCACATAAACAGAGGCATCCTATTTGAATTCATGCATCATTTTA
GCAGGAACCTATAGAGCATTCAGAAAGTAAATGATCTGTATGAAGATTTGGTTTTTCACTGTCCATCCCT
ATGGTGGATGAGTTCATAAATCCTCTAGTCTTTTACCTCATTATAAATTAGCCTTTGGATTTTGTGAGCG
CACATCATTTAGCCTGTGAACTGTACTCAGGGGATCCATAAACGTTCCCGCTGCAGACTCAGATCTTGAT
TTATTGTTTTGAT
CCGGAACACAACTGTTGCACTCTATTTCACATAAACAGAGGCATCCTATTTGAATTCATGCATCATTTTA
GCAGGAACCTATAGAGCATTCAGAAAGTAAATGATCTGTATGAAGATTTGGTTTTTCACTGTCCATCCCT
ATGGTGGATGAGTTCATAAATCCTCTAGTCTTTTACCTCATTATAAATTAGCCTTTGGATTTTGTGAGCG
CACATCATTTAGCCTGTGAACTGTACTCAGGGGATCCATAAACGTTCCCGCTGCAGACTCAGATCTTGAT
TTATTGTTTTGAT