ZFLNCG05837
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR941749 | anterior pectoral fin | normal | 175.59 |
| SRR941753 | posterior pectoral fin | normal | 174.76 |
| SRR1205160 | 5dpf | transgenic sqET20 and GFP+ | 128.51 |
| SRR592703 | pineal gland | normal | 89.15 |
| SRR592702 | pineal gland | normal | 86.81 |
| SRR1205154 | 5dpf | transgenic sqET20 | 81.65 |
| ERR023144 | brain | normal | 76.46 |
| SRR1028002 | head | normal | 73.30 |
| SRR1205157 | 5dpf | transgenic sqET20 and neomycin treated 1h and GFP+ | 72.36 |
| SRR527834 | head | normal | 71.24 |
Express in tissues
Correlated coding gene
Download
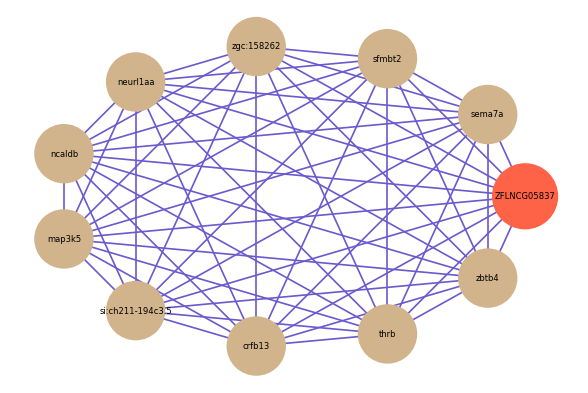
| gene | correlation coefficent |
|---|---|
| sema7a | 0.78 |
| sfmbt2 | 0.78 |
| zgc:158262 | 0.78 |
| neurl1aa | 0.77 |
| ncaldb | 0.77 |
| map3k5 | 0.77 |
| si:ch211-194c3.5 | 0.76 |
| crfb13 | 0.75 |
| thrb | 0.75 |
| zbtb4 | 0.75 |
Gene Ontology
Download
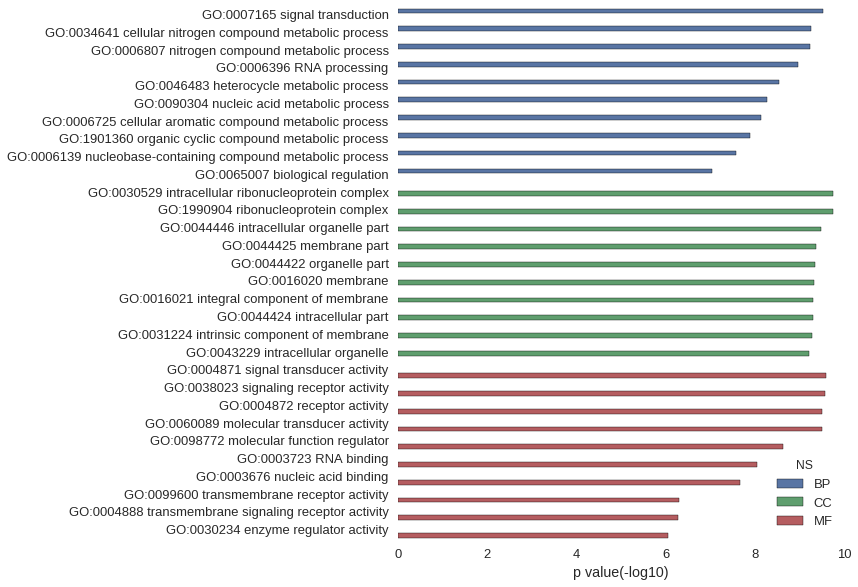
| GO | P value |
|---|---|
| GO:0007165 | 3.00e-10 |
| GO:0034641 | 5.66e-10 |
| GO:0006807 | 5.73e-10 |
| GO:0006396 | 1.09e-09 |
| GO:0046483 | 2.91e-09 |
| GO:0090304 | 5.45e-09 |
| GO:0006725 | 7.23e-09 |
| GO:1901360 | 1.32e-08 |
| GO:0006139 | 2.60e-08 |
| GO:0065007 | 9.10e-08 |
| GO:0030529 | 1.82e-10 |
| GO:1990904 | 1.82e-10 |
| GO:0044446 | 3.37e-10 |
| GO:0044425 | 4.23e-10 |
| GO:0044422 | 4.56e-10 |
| GO:0016020 | 4.66e-10 |
| GO:0016021 | 5.00e-10 |
| GO:0044424 | 5.05e-10 |
| GO:0031224 | 5.40e-10 |
| GO:0043229 | 6.01e-10 |
| GO:0004871 | 2.57e-10 |
| GO:0038023 | 2.71e-10 |
| GO:0004872 | 3.09e-10 |
| GO:0060089 | 3.09e-10 |
| GO:0098772 | 2.29e-09 |
| GO:0003723 | 8.85e-09 |
| GO:0003676 | 2.17e-08 |
| GO:0099600 | 5.04e-07 |
| GO:0004888 | 5.40e-07 |
| GO:0030234 | 8.92e-07 |
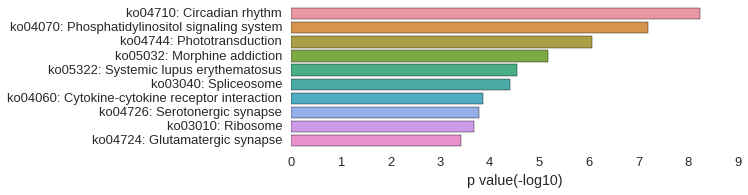
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG05837
ACAGAACAGAACTGTTCAAGTATTTTACACTTATTTTATCCTTAAATGAAAGTTTTAAGAAAAATTCAAA
GACTTCGCTGAGCACATATCAGGGCCTTTACAGATCAGTCCAGGGAGCAAGCAGTTGGGTGGTGTTCGAA
GGGTTAAGCAGCAAAAAAGGCCGGTCTGCACCGGATCGTCATGGGCAGAGAACATAAATACTGATTGGCA
TGGGCAAATAAAATAAGCAGAGGAGAATGGTAGTGTGAAATTTACCATCAGCAAACGGGTCATGACGCTC
TGGGGAGATGATCATCTTTTGTCTGCGCTTCTTGTATTCATCCTCCCTGTCTGAAATTTTCTGCGGTCTG
TGTTCGGCAAAGGGATCATACTGCAAAATGGGTACAAACAATTGTATGATCAATTCGAAATTGACATTTC
AAAAACACATACAAAAACTTCACAAGAAAACACACACACACACACTGCAACTTGAAGGTGGAAAGTCTGT
TAAATTACCTGCTCATCTGACTGAGGAATTGAGTTGAGTATTGCAACCGGAGCATGATATCCTGGTTTCT
TCTGTACAAGGAGACTTGTAGATGAATCCTCGTCATCATCCTAAAATAAAAATAGTTGGGGTTAGGTTCA
AGAAACTAAACAGAATTCTTATACACAATGGAGTTGTATTTTTTGTTTTACTTTTAAGAGCGTTATGCTA
TAACTTACATCCTCCTGCTCATTGGCAGCAATTGAAGTCAAATACCCATCAAAGCGGCTGTCACTGCCCC
CGTAGATCTCCTGGTCATAATACCCAGTGGAATCAAGGCCAACTCCTTGGTCACCGCCCTCCACCACCAG
AGCGGCTTTCTTCCCCTGGATTTCCAGGATCTGAGCCTCAATGT
ACAGAACAGAACTGTTCAAGTATTTTACACTTATTTTATCCTTAAATGAAAGTTTTAAGAAAAATTCAAA
GACTTCGCTGAGCACATATCAGGGCCTTTACAGATCAGTCCAGGGAGCAAGCAGTTGGGTGGTGTTCGAA
GGGTTAAGCAGCAAAAAAGGCCGGTCTGCACCGGATCGTCATGGGCAGAGAACATAAATACTGATTGGCA
TGGGCAAATAAAATAAGCAGAGGAGAATGGTAGTGTGAAATTTACCATCAGCAAACGGGTCATGACGCTC
TGGGGAGATGATCATCTTTTGTCTGCGCTTCTTGTATTCATCCTCCCTGTCTGAAATTTTCTGCGGTCTG
TGTTCGGCAAAGGGATCATACTGCAAAATGGGTACAAACAATTGTATGATCAATTCGAAATTGACATTTC
AAAAACACATACAAAAACTTCACAAGAAAACACACACACACACACTGCAACTTGAAGGTGGAAAGTCTGT
TAAATTACCTGCTCATCTGACTGAGGAATTGAGTTGAGTATTGCAACCGGAGCATGATATCCTGGTTTCT
TCTGTACAAGGAGACTTGTAGATGAATCCTCGTCATCATCCTAAAATAAAAATAGTTGGGGTTAGGTTCA
AGAAACTAAACAGAATTCTTATACACAATGGAGTTGTATTTTTTGTTTTACTTTTAAGAGCGTTATGCTA
TAACTTACATCCTCCTGCTCATTGGCAGCAATTGAAGTCAAATACCCATCAAAGCGGCTGTCACTGCCCC
CGTAGATCTCCTGGTCATAATACCCAGTGGAATCAAGGCCAACTCCTTGGTCACCGCCCTCCACCACCAG
AGCGGCTTTCTTCCCCTGGATTTCCAGGATCTGAGCCTCAATGT