ZFLNCG06016
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR023143 | swim bladder | normal | 201.87 |
| ERR023146 | head kidney | normal | 168.32 |
| ERR023145 | heart | normal | 142.93 |
| SRR1647679 | head kidney | normal | 66.45 |
| SRR1299124 | caudal fin | Zero day time after treatment | 58.30 |
| SRR1647680 | head kidney | SVCV treatment | 55.78 |
| SRR592700 | pineal gland | normal | 54.50 |
| SRR592702 | pineal gland | normal | 54.33 |
| SRR1647681 | head kidney | SVCV treatment | 48.36 |
| ERR023144 | brain | normal | 47.76 |
Express in tissues
Correlated coding gene
Download
Gene Ontology
Download
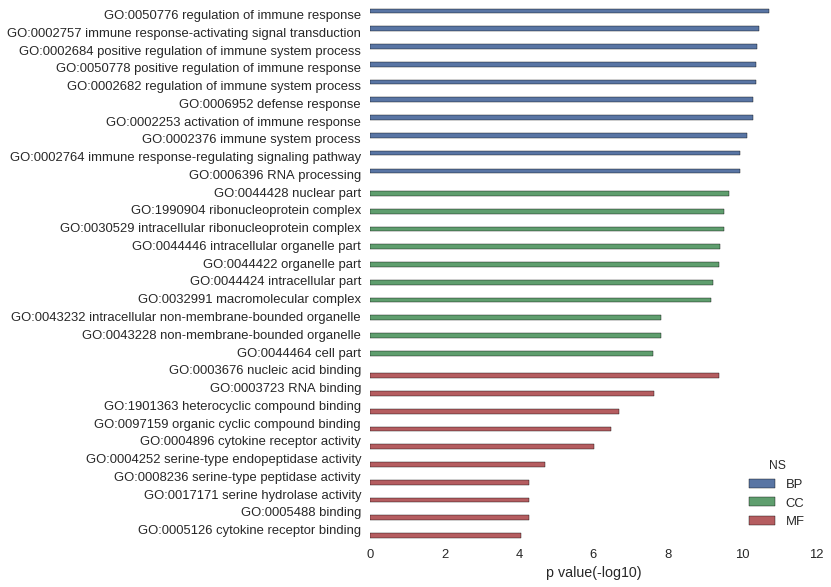
| GO | P value |
|---|---|
| GO:0050776 | 1.85e-11 |
| GO:0002757 | 3.52e-11 |
| GO:0002684 | 4.03e-11 |
| GO:0050778 | 4.17e-11 |
| GO:0002682 | 4.30e-11 |
| GO:0006952 | 5.06e-11 |
| GO:0002253 | 5.16e-11 |
| GO:0002376 | 7.53e-11 |
| GO:0002764 | 1.17e-10 |
| GO:0006396 | 1.18e-10 |
| GO:0044428 | 2.32e-10 |
| GO:1990904 | 3.04e-10 |
| GO:0030529 | 3.04e-10 |
| GO:0044446 | 4.05e-10 |
| GO:0044422 | 4.19e-10 |
| GO:0044424 | 6.01e-10 |
| GO:0032991 | 6.91e-10 |
| GO:0043232 | 1.48e-08 |
| GO:0043228 | 1.48e-08 |
| GO:0044464 | 2.49e-08 |
| GO:0003676 | 4.27e-10 |
| GO:0003723 | 2.41e-08 |
| GO:1901363 | 2.08e-07 |
| GO:0097159 | 3.26e-07 |
| GO:0004896 | 9.76e-07 |
| GO:0004252 | 1.98e-05 |
| GO:0008236 | 5.28e-05 |
| GO:0017171 | 5.28e-05 |
| GO:0005488 | 5.38e-05 |
| GO:0005126 | 9.02e-05 |
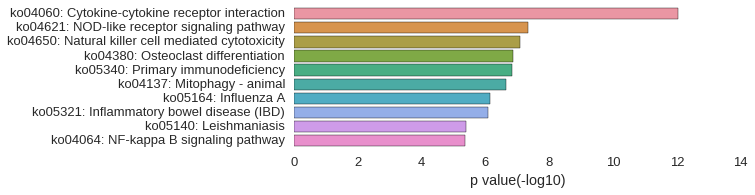
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG06016
ATATTTTTATGTATAGTTATAATTAAACTACAGACACCACTAACAAGCGCTGAAGAGGGCAAAATGCCAT
ATATTTTGTGCACTATATATTTTAAAAGCGCCTTAAGGTCATATTTAGTTTAATCTTTCTGTGCAATCAA
TCAGAGCTTATTGAGTGCCAAATTTTGCGTCCGCTTCTGCTGTGAGATATTTTACCACTAACAGATTATG
AGGACCAGTGTTTCTTTTCTGCCTTTCTCTGCTGAAAAGGTCATTTTTCTTTAGTTAGTTTGTTAGATAT
GCACATGTCCTGATTGTGATCCTGATGTTTTTAAATGTTTTATGTTGTTGTTGGTGATTTTCTGATTCGT
CTTTGCCACTAACAGTTATCTGTATCTATATTGTTATGAATTAAACTGTATAAACATTGATTAAGGCTTT
ATTTATCCATTCATGAACAAGCCATTTCTCTGAGAGAAACTAATCGTGTTGCTCTTTCAATACAAATAAA
GATTCG
ATATTTTTATGTATAGTTATAATTAAACTACAGACACCACTAACAAGCGCTGAAGAGGGCAAAATGCCAT
ATATTTTGTGCACTATATATTTTAAAAGCGCCTTAAGGTCATATTTAGTTTAATCTTTCTGTGCAATCAA
TCAGAGCTTATTGAGTGCCAAATTTTGCGTCCGCTTCTGCTGTGAGATATTTTACCACTAACAGATTATG
AGGACCAGTGTTTCTTTTCTGCCTTTCTCTGCTGAAAAGGTCATTTTTCTTTAGTTAGTTTGTTAGATAT
GCACATGTCCTGATTGTGATCCTGATGTTTTTAAATGTTTTATGTTGTTGTTGGTGATTTTCTGATTCGT
CTTTGCCACTAACAGTTATCTGTATCTATATTGTTATGAATTAAACTGTATAAACATTGATTAAGGCTTT
ATTTATCCATTCATGAACAAGCCATTTCTCTGAGAGAAACTAATCGTGTTGCTCTTTCAATACAAATAAA
GATTCG