ZFLNCG06179
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1562529 | intestine and pancreas | normal | 26.54 |
| SRR941753 | posterior pectoral fin | normal | 25.45 |
| SRR516123 | skin | male and 3.5 year | 24.50 |
| SRR516121 | skin | male and 3.5 year | 20.53 |
| ERR023146 | head kidney | normal | 17.08 |
| SRR516122 | skin | male and 3.5 year | 16.99 |
| SRR941749 | anterior pectoral fin | normal | 16.28 |
| SRR516129 | skin | male and 5 month | 16.10 |
| SRR592703 | pineal gland | normal | 15.99 |
| SRR1562531 | muscle | normal | 14.74 |
Express in tissues
Correlated coding gene
Download
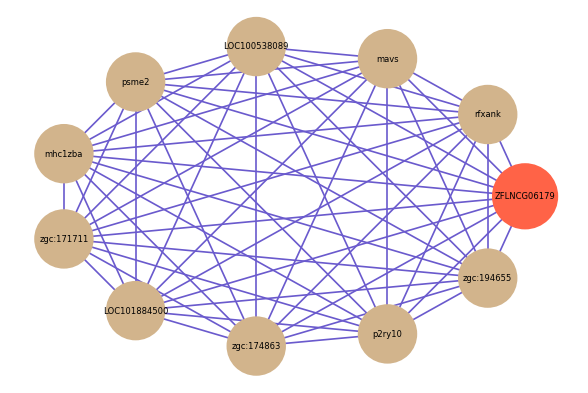
| gene | correlation coefficent |
|---|---|
| rfxank | 0.69 |
| mavs | 0.68 |
| LOC100538089 | 0.68 |
| psme2 | 0.67 |
| mhc1zba | 0.67 |
| zgc:171711 | 0.67 |
| LOC101884500 | 0.67 |
| zgc:174863 | 0.67 |
| p2ry10 | 0.66 |
| zgc:194655 | 0.66 |
Gene Ontology
Download
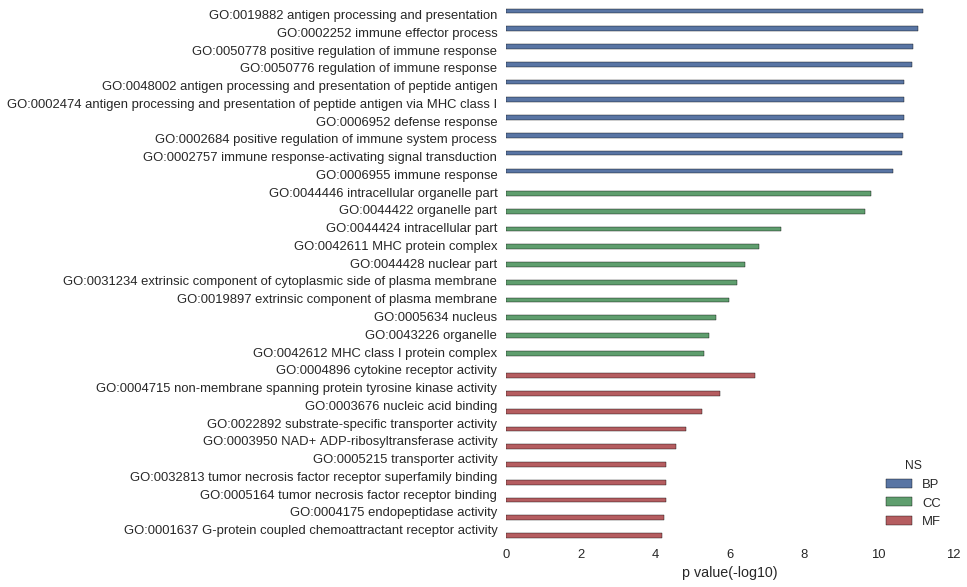
| GO | P value |
|---|---|
| GO:0019882 | 6.20e-12 |
| GO:0002252 | 8.45e-12 |
| GO:0050778 | 1.19e-11 |
| GO:0050776 | 1.24e-11 |
| GO:0048002 | 2.02e-11 |
| GO:0002474 | 2.02e-11 |
| GO:0006952 | 2.11e-11 |
| GO:0002684 | 2.12e-11 |
| GO:0002757 | 2.33e-11 |
| GO:0006955 | 4.11e-11 |
| GO:0044446 | 1.63e-10 |
| GO:0044422 | 2.32e-10 |
| GO:0044424 | 4.11e-08 |
| GO:0042611 | 1.66e-07 |
| GO:0044428 | 3.94e-07 |
| GO:0031234 | 6.17e-07 |
| GO:0019897 | 1.07e-06 |
| GO:0005634 | 2.38e-06 |
| GO:0043226 | 3.52e-06 |
| GO:0042612 | 4.99e-06 |
| GO:0004896 | 2.10e-07 |
| GO:0004715 | 1.78e-06 |
| GO:0003676 | 5.61e-06 |
| GO:0022892 | 1.51e-05 |
| GO:0003950 | 2.70e-05 |
| GO:0005215 | 5.03e-05 |
| GO:0032813 | 5.10e-05 |
| GO:0005164 | 5.10e-05 |
| GO:0004175 | 5.87e-05 |
| GO:0001637 | 6.47e-05 |
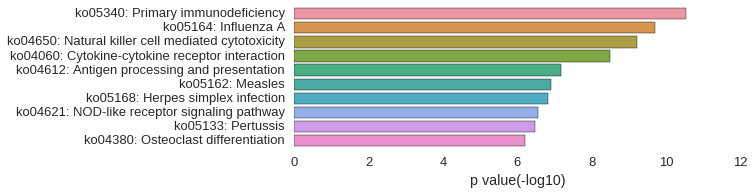
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG06179
GAAGATGATAGGAACACTTCTTCACCCATATACGAGAATACAGAAAGTACCCCGCTGATTCACAGTGACG
ACTCAAGTGAAGGAGTCATACAAAGACAAGAAGATGTACCAAGACATGAAGACAAACTAAGACAAGCAGA
CAAACTAAGACAAGATGCTTGTTGTTTTTGTGGGGTTTGCTTCTTCTTTTGTTGTGGTTGGCTTTGTGAA
TGTAAACCTTATA
GAAGATGATAGGAACACTTCTTCACCCATATACGAGAATACAGAAAGTACCCCGCTGATTCACAGTGACG
ACTCAAGTGAAGGAGTCATACAAAGACAAGAAGATGTACCAAGACATGAAGACAAACTAAGACAAGCAGA
CAAACTAAGACAAGATGCTTGTTGTTTTTGTGGGGTTTGCTTCTTCTTTTGTTGTGGTTGGCTTTGTGAA
TGTAAACCTTATA