ZFLNCG06713
Basic Information
Chromesome: chr11
Start: 12695339
End: 12695633
Transcript: ZFLNCT10276
Known as: ENSDARG00000088209
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR800046 | sphere stage | 5azaCyD treatment | 240.16 |
| SRR800037 | egg | normal | 230.49 |
| SRR800049 | sphere stage | control treatment | 220.97 |
| SRR800043 | sphere stage | normal | 184.57 |
| SRR748488 | sperm | normal | 32.02 |
| SRR800045 | muscle | normal | 15.84 |
| SRR372796 | bud | normal | 11.50 |
| SRR801555 | embryo | RPS19 morpholino | 9.97 |
| SRR801556 | embryo | RPS19 and p53 morpholino | 9.91 |
| SRR372791 | dome | normal | 8.81 |
Express in tissues
Correlated coding gene
Download
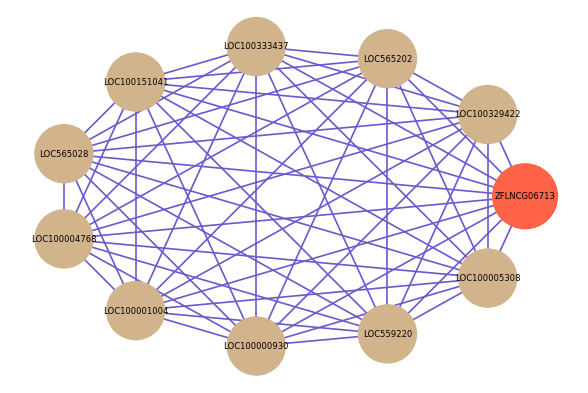
| gene | correlation coefficent |
|---|---|
| LOC100329422 | 0.76 |
| LOC565202 | 0.72 |
| LOC100333437 | 0.71 |
| LOC100151041 | 0.69 |
| LOC565028 | 0.69 |
| LOC100004768 | 0.68 |
| LOC100001004 | 0.68 |
| LOC100000930 | 0.68 |
| LOC559220 | 0.67 |
| LOC100005308 | 0.67 |
Gene Ontology
Download
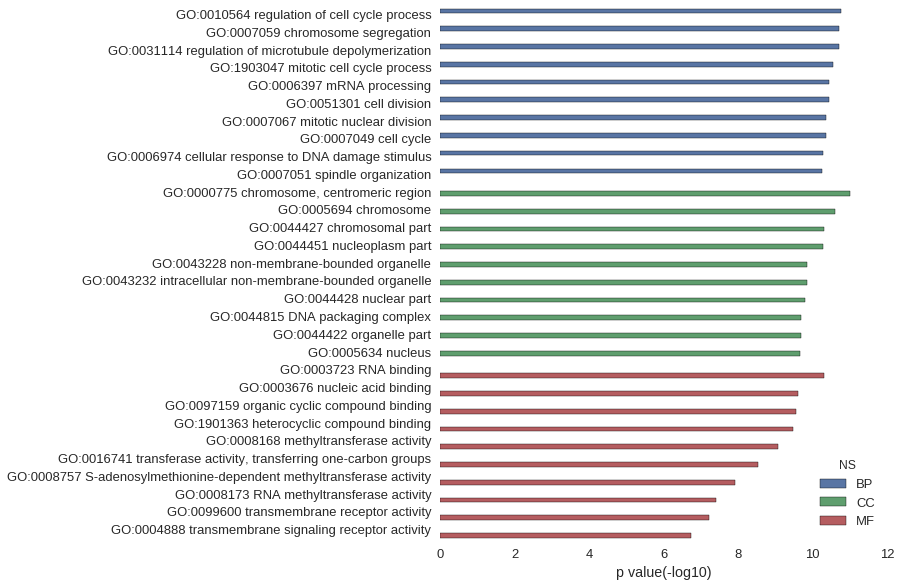
| GO | P value |
|---|---|
| GO:0010564 | 1.67e-11 |
| GO:0007059 | 1.93e-11 |
| GO:0031114 | 1.93e-11 |
| GO:1903047 | 2.85e-11 |
| GO:0006397 | 3.58e-11 |
| GO:0051301 | 3.66e-11 |
| GO:0007067 | 4.37e-11 |
| GO:0007049 | 4.44e-11 |
| GO:0006974 | 5.07e-11 |
| GO:0007051 | 5.59e-11 |
| GO:0000775 | 9.64e-12 |
| GO:0005694 | 2.48e-11 |
| GO:0044427 | 4.80e-11 |
| GO:0044451 | 5.07e-11 |
| GO:0043228 | 1.43e-10 |
| GO:0043232 | 1.43e-10 |
| GO:0044428 | 1.58e-10 |
| GO:0044815 | 2.02e-10 |
| GO:0044422 | 2.05e-10 |
| GO:0005634 | 2.16e-10 |
| GO:0003723 | 4.77e-11 |
| GO:0003676 | 2.45e-10 |
| GO:0097159 | 2.69e-10 |
| GO:1901363 | 3.30e-10 |
| GO:0008168 | 8.46e-10 |
| GO:0016741 | 2.89e-09 |
| GO:0008757 | 1.24e-08 |
| GO:0008173 | 3.84e-08 |
| GO:0099600 | 6.09e-08 |
| GO:0004888 | 1.87e-07 |
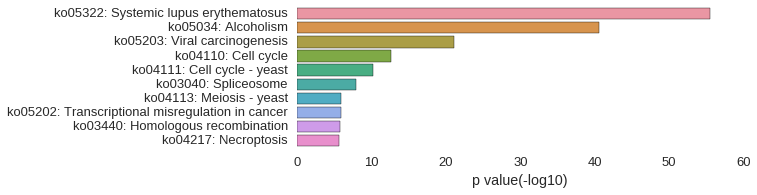
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG06713
GTCTGCGTCCCTCTGTGTCGCTCAGGCTGCACTGCAGTGTCTATTCACAGGCGCGATCCCACTACTGATC
AGCACGGGGGATTTGACCTGCTCCGTCTCCGACCTGGGCCGGTTCACCCCTCCTTAAACGACCTGGTGGT
CCCGAGCTCCCCCAGGAGAACCATATCGATACCGAACTTAGCGCGGACACCCGATCAACATAGTCCACTG
CAGCCCAGAAGCCCTGAGCTCAAGCGATCCGCAGCCTCAGCCTCCCTGTAGCTTGGATTACAGGCGCGCG
CCACTGAACCCGGC
GTCTGCGTCCCTCTGTGTCGCTCAGGCTGCACTGCAGTGTCTATTCACAGGCGCGATCCCACTACTGATC
AGCACGGGGGATTTGACCTGCTCCGTCTCCGACCTGGGCCGGTTCACCCCTCCTTAAACGACCTGGTGGT
CCCGAGCTCCCCCAGGAGAACCATATCGATACCGAACTTAGCGCGGACACCCGATCAACATAGTCCACTG
CAGCCCAGAAGCCCTGAGCTCAAGCGATCCGCAGCCTCAGCCTCCCTGTAGCTTGGATTACAGGCGCGCG
CCACTGAACCCGGC