ZFLNCG06721
Basic Information
Chromesome: chr11
Start: 12769632
End: 12769926
Transcript: ZFLNCT10284
Known as: ENSDARG00000089521
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR800037 | egg | normal | 3174.42 |
| SRR800046 | sphere stage | 5azaCyD treatment | 2716.62 |
| SRR800049 | sphere stage | control treatment | 1934.06 |
| SRR800043 | sphere stage | normal | 1624.93 |
| SRR886457 | 512 cell | 4-thio-UTP metabolic labeling | 303.76 |
| SRR800045 | muscle | normal | 118.81 |
| SRR886455 | 128 cell | 4-thio-UTP metabolic labeling | 87.20 |
| SRR886456 | 256 cell | 4-thio-UTP metabolic labeling | 69.22 |
| SRR748488 | sperm | normal | 42.14 |
| SRR801555 | embryo | RPS19 morpholino | 25.48 |
Express in tissues
Correlated coding gene
Download
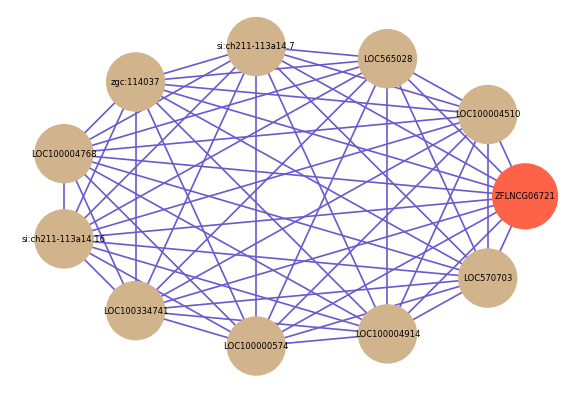
| gene | correlation coefficent |
|---|---|
| LOC100004510 | 0.74 |
| LOC565028 | 0.71 |
| si:ch211-113a14.7 | 0.70 |
| zgc:114037 | 0.70 |
| LOC100004768 | 0.70 |
| si:ch211-113a14.16 | 0.70 |
| LOC100334741 | 0.70 |
| LOC100000574 | 0.69 |
| LOC100004914 | 0.69 |
| LOC570703 | 0.69 |
Gene Ontology
Download
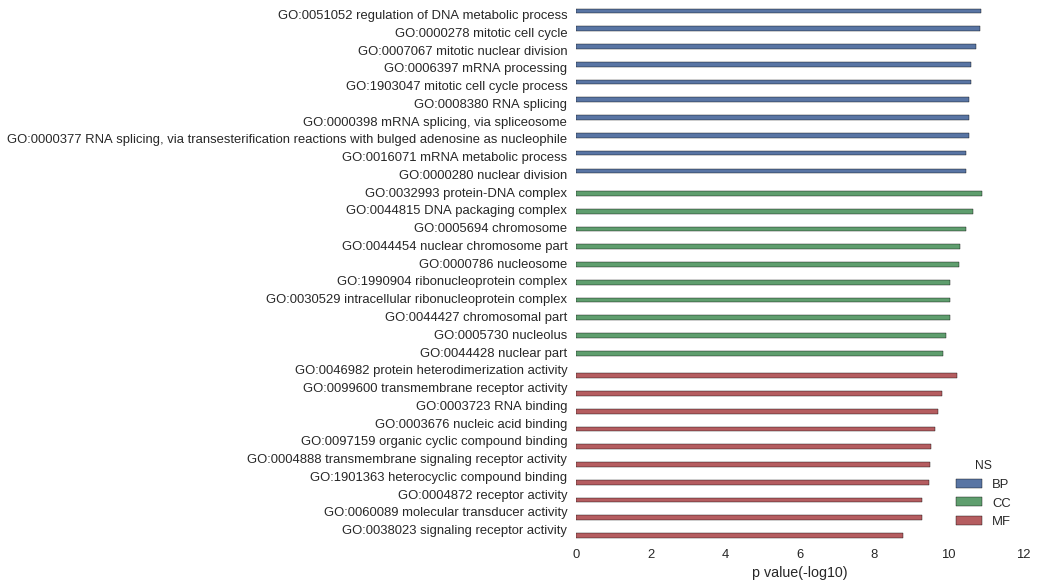
| GO | P value |
|---|---|
| GO:0051052 | 1.31e-11 |
| GO:0000278 | 1.43e-11 |
| GO:0007067 | 1.82e-11 |
| GO:0006397 | 2.43e-11 |
| GO:1903047 | 2.47e-11 |
| GO:0008380 | 2.73e-11 |
| GO:0000398 | 2.77e-11 |
| GO:0000377 | 2.77e-11 |
| GO:0016071 | 3.36e-11 |
| GO:0000280 | 3.39e-11 |
| GO:0032993 | 1.27e-11 |
| GO:0044815 | 2.14e-11 |
| GO:0005694 | 3.28e-11 |
| GO:0044454 | 4.75e-11 |
| GO:0000786 | 5.22e-11 |
| GO:1990904 | 8.83e-11 |
| GO:0030529 | 8.83e-11 |
| GO:0044427 | 8.99e-11 |
| GO:0005730 | 1.13e-10 |
| GO:0044428 | 1.38e-10 |
| GO:0046982 | 5.93e-11 |
| GO:0099600 | 1.51e-10 |
| GO:0003723 | 1.89e-10 |
| GO:0003676 | 2.28e-10 |
| GO:0097159 | 2.90e-10 |
| GO:0004888 | 3.15e-10 |
| GO:1901363 | 3.42e-10 |
| GO:0004872 | 5.27e-10 |
| GO:0060089 | 5.27e-10 |
| GO:0038023 | 1.62e-09 |
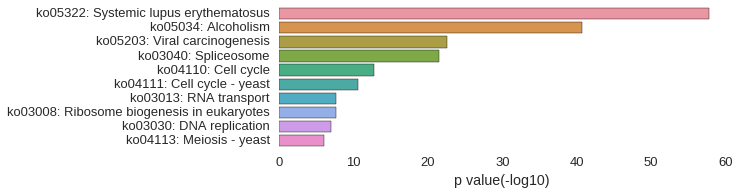
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG06721
CCGGGTTCAGTGGCGCGCGCCTGTAATCCAAGCTACTGGGAGGCTGAGGCTGCGGATCGCTTGAGCTCAG
GGCTTCTGGGCTGCAGTGGACTATGTTGATCGGGTGTCTGCGCTAAGTTCGGTATCGATATGGTTCTCCT
GGGGGAGCTCGGGACCACCAGGTCGTTTAAGGAGGGGTGAACCGGCCCAGGTCGGAGACGGAGCAGGTCA
AATCCCCCGTGCTGATCAGTAGTGGGATCGCGCCTGTGAATAGACACTGCAGTGCAGCCTGAGCGACAAA
GAGAGACGCAGACT
CCGGGTTCAGTGGCGCGCGCCTGTAATCCAAGCTACTGGGAGGCTGAGGCTGCGGATCGCTTGAGCTCAG
GGCTTCTGGGCTGCAGTGGACTATGTTGATCGGGTGTCTGCGCTAAGTTCGGTATCGATATGGTTCTCCT
GGGGGAGCTCGGGACCACCAGGTCGTTTAAGGAGGGGTGAACCGGCCCAGGTCGGAGACGGAGCAGGTCA
AATCCCCCGTGCTGATCAGTAGTGGGATCGCGCCTGTGAATAGACACTGCAGTGCAGCCTGAGCGACAAA
GAGAGACGCAGACT