ZFLNCG06844
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR023144 | brain | normal | 61.91 |
| SRR1565811 | brain | normal | 21.69 |
| SRR1565809 | brain | normal | 18.90 |
| SRR1565807 | brain | normal | 18.12 |
| SRR1565819 | brain | normal | 18.05 |
| SRR1565817 | brain | normal | 17.60 |
| SRR1565805 | brain | normal | 16.03 |
| SRR1565815 | brain | normal | 13.77 |
| SRR726542 | 5 dpf | infection with control | 10.80 |
| SRR1565813 | brain | normal | 9.73 |
Express in tissues
Correlated coding gene
Download
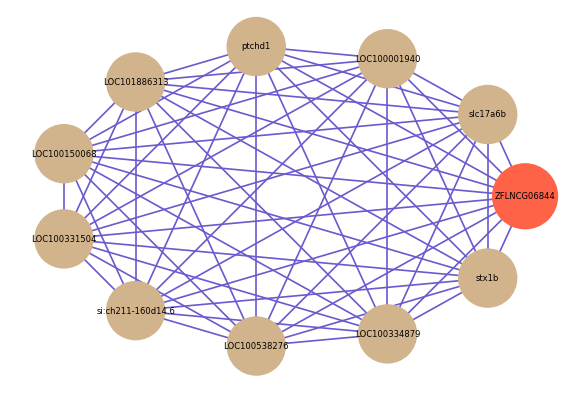
| gene | correlation coefficent |
|---|---|
| slc17a6b | 0.83 |
| LOC100001940 | 0.82 |
| ptchd1 | 0.82 |
| LOC101886313 | 0.82 |
| LOC100150068 | 0.82 |
| LOC100331504 | 0.82 |
| si:ch211-160d14.6 | 0.82 |
| LOC100538276 | 0.81 |
| LOC100334879 | 0.81 |
| stx1b | 0.81 |
Gene Ontology
Download
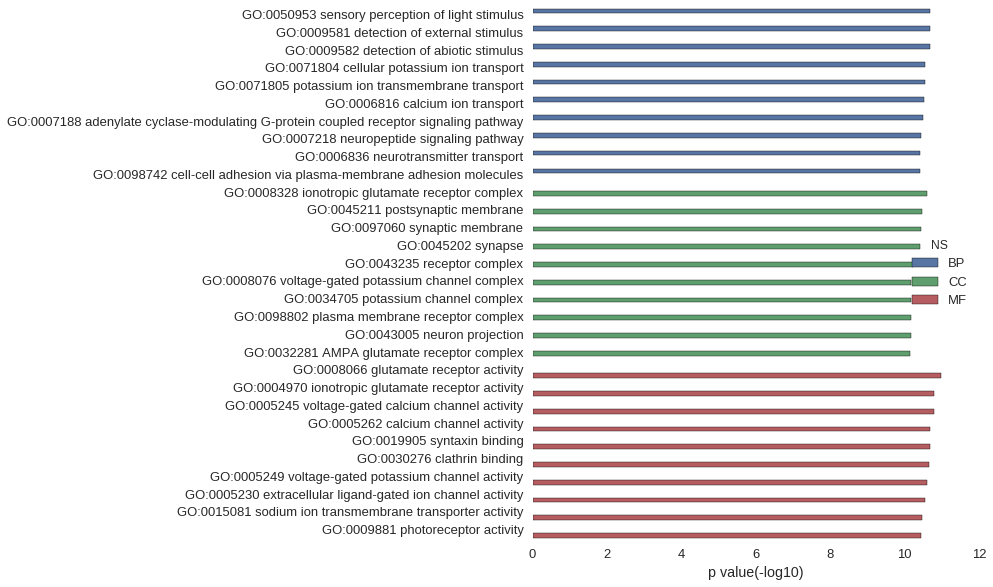
| GO | P value |
|---|---|
| GO:0050953 | 2.03e-11 |
| GO:0009581 | 2.06e-11 |
| GO:0009582 | 2.06e-11 |
| GO:0071804 | 2.82e-11 |
| GO:0071805 | 2.82e-11 |
| GO:0006816 | 3.05e-11 |
| GO:0007188 | 3.15e-11 |
| GO:0007218 | 3.69e-11 |
| GO:0006836 | 3.90e-11 |
| GO:0098742 | 3.92e-11 |
| GO:0008328 | 2.46e-11 |
| GO:0045211 | 3.45e-11 |
| GO:0097060 | 3.59e-11 |
| GO:0045202 | 3.92e-11 |
| GO:0043235 | 6.03e-11 |
| GO:0008076 | 6.84e-11 |
| GO:0034705 | 6.84e-11 |
| GO:0098802 | 6.90e-11 |
| GO:0043005 | 6.90e-11 |
| GO:0032281 | 7.36e-11 |
| GO:0008066 | 1.04e-11 |
| GO:0004970 | 1.57e-11 |
| GO:0005245 | 1.58e-11 |
| GO:0005262 | 2.03e-11 |
| GO:0019905 | 2.06e-11 |
| GO:0030276 | 2.22e-11 |
| GO:0005249 | 2.49e-11 |
| GO:0005230 | 2.82e-11 |
| GO:0015081 | 3.44e-11 |
| GO:0009881 | 3.68e-11 |
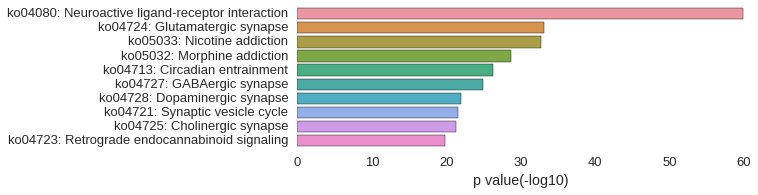
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG06844
GAATATTTTGTCAATTCAAATGACTAGCGGCTTCGACCTGGAAACAGTATTTCATACGTCACGGCTTAAC
AAGCGGATGGGTTTACTCAGCAATGTTTACTAATCACTTTTCTGGATTTTGACTGTGGTTTGCATATCTT
AGAATGCTCAGCACTAAACATTAAAGCTATTTAGATGAGCCTGGTCACTCGCCAGATGTTGCTTTTACAG
CTGGTTAGAACAAAAAACTCAATGTAATCATGCGTCACATCACGTCTGGGTAGGACACCGTGTGTTTTAT
GCAGTTGTTTTTGTATCTCGGTTGCATGTGTGTTGCATTTCTG
GAATATTTTGTCAATTCAAATGACTAGCGGCTTCGACCTGGAAACAGTATTTCATACGTCACGGCTTAAC
AAGCGGATGGGTTTACTCAGCAATGTTTACTAATCACTTTTCTGGATTTTGACTGTGGTTTGCATATCTT
AGAATGCTCAGCACTAAACATTAAAGCTATTTAGATGAGCCTGGTCACTCGCCAGATGTTGCTTTTACAG
CTGGTTAGAACAAAAAACTCAATGTAATCATGCGTCACATCACGTCTGGGTAGGACACCGTGTGTTTTAT
GCAGTTGTTTTTGTATCTCGGTTGCATGTGTGTTGCATTTCTG