ZFLNCG06985
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR700534 | heart | control morpholino | 14.90 |
| SRR891511 | brain | normal | 12.68 |
| SRR592698 | pineal gland | normal | 5.98 |
| SRR519727 | 6 dpf | FETOH treatment | 5.96 |
| SRR519732 | 7 dpf | FETOH treatment | 5.54 |
| SRR519747 | 6 dpf | VD3 treatment | 5.19 |
| SRR592700 | pineal gland | normal | 5.05 |
| SRR700537 | heart | Gata4 morpholino | 5.02 |
| SRR1565817 | brain | normal | 4.96 |
| SRR1565819 | brain | normal | 4.93 |
Express in tissues
Correlated coding gene
Download
Gene Ontology
Download
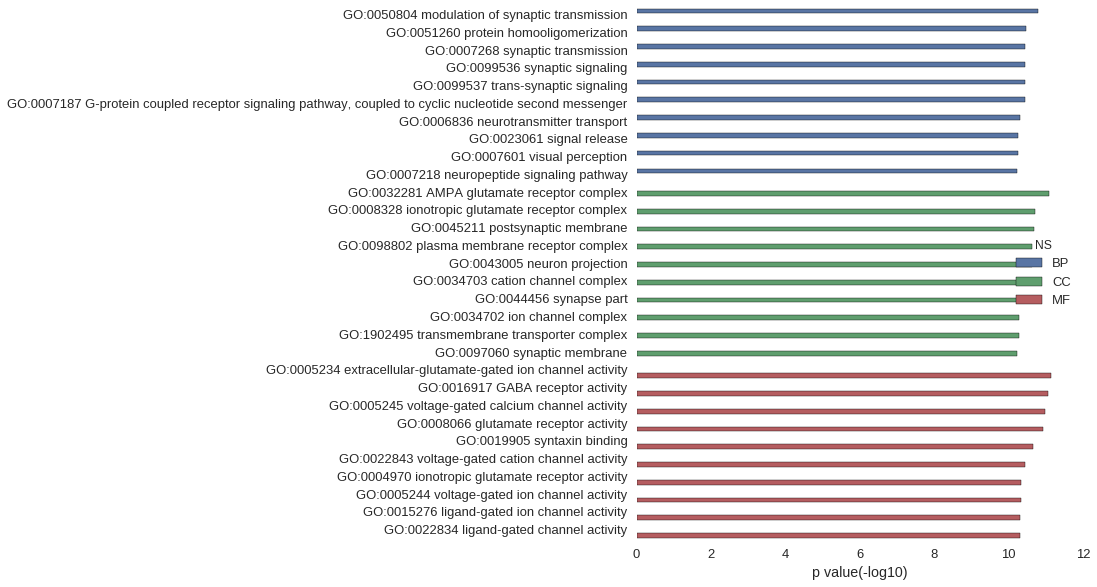
| GO | P value |
|---|---|
| GO:0050804 | 1.65e-11 |
| GO:0051260 | 3.39e-11 |
| GO:0007268 | 3.50e-11 |
| GO:0099536 | 3.50e-11 |
| GO:0099537 | 3.50e-11 |
| GO:0007187 | 3.66e-11 |
| GO:0006836 | 4.89e-11 |
| GO:0023061 | 5.64e-11 |
| GO:0007601 | 5.68e-11 |
| GO:0007218 | 5.77e-11 |
| GO:0032281 | 8.06e-12 |
| GO:0008328 | 1.97e-11 |
| GO:0045211 | 2.01e-11 |
| GO:0098802 | 2.30e-11 |
| GO:0043005 | 2.30e-11 |
| GO:0034703 | 3.13e-11 |
| GO:0044456 | 4.73e-11 |
| GO:0034702 | 5.17e-11 |
| GO:1902495 | 5.23e-11 |
| GO:0097060 | 5.76e-11 |
| GO:0005234 | 7.11e-12 |
| GO:0016917 | 8.55e-12 |
| GO:0005245 | 1.03e-11 |
| GO:0008066 | 1.17e-11 |
| GO:0019905 | 2.22e-11 |
| GO:0022843 | 3.55e-11 |
| GO:0004970 | 4.49e-11 |
| GO:0005244 | 4.73e-11 |
| GO:0015276 | 4.83e-11 |
| GO:0022834 | 4.83e-11 |
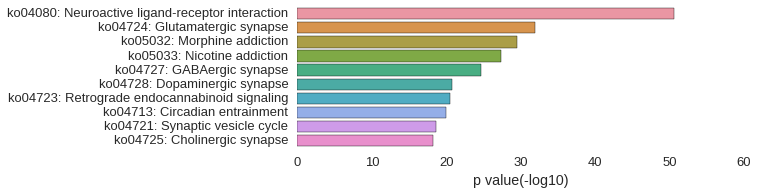
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG06985
TGACGCTCTGATCAACCTGTGCACTCTCACACACACACACACACACACCGTACTCTGTATAAATGAAGCA
TCACACACTCACACACACACACACACACACACACACACACACACACACACACACACACACACACACCGTA
CTCTGTATAAATGAAGCATCACACACTCACACACACACACACACACACACACACACACACACACACACAC
ACCGTACTCTGTATAAATGAAGCATCACACACACACACACTCACACACACACACACACACACACACACAA
ACACACACACACACACACACACACACACACACACACACACACACACACACACACCATACTCTGTATAAAT
GAAGCATCACACACACACACACTCACACACACACACACACACACACACACACACACACACACACACACAC
ACACACCATACTCTGTATAAATGAAGCATCACACACACACACACACATACTCCATACTCTGAATTAATGA
AGTGTAATGTTTCCTCTACCAGATGTTTCCGAACTTCTCCAAGGACTCCACCAGATCAGCCTCCACCACA
GACTCACAGAGACCCCGAACATGGACCACAGGAGACGGAGCGATTCTGTGTGTCTCTGAGCTGCTGTCCT
GAGGAGGAAGACATGACAGATCAACACTACACTCACTCATTCATTCATTCATTCATTCATTCATTCATTC
ATTCATTCATTCATTCATTCATTCATTCACTCATTCATTCATTCATTGACAGAGCAATTCTGTGCATCTC
TGAGCAGCTGTCCTGAAGAGACATGACAGATCAACACCACATTCATTCATTCATTCATTCATTCATTCAT
TCACTCATTCATTCATTCATTGACAGAGCAATTCTGTGCATCTCTAAGCAGCTGTCTTGAAGAGACATGA
CAGATCAACACCACATTCATTTATTAATTAATTAATTCATTCATTCATTCCTTCATTTATCAGTTCCTTC
AGTCATTCATTAATTCAGTTATTAAATCATCAACAGGGCTATTTTGTGTGTCTCTGAGCGCTATCCTGAG
GACGGA
TGACGCTCTGATCAACCTGTGCACTCTCACACACACACACACACACACCGTACTCTGTATAAATGAAGCA
TCACACACTCACACACACACACACACACACACACACACACACACACACACACACACACACACACACCGTA
CTCTGTATAAATGAAGCATCACACACTCACACACACACACACACACACACACACACACACACACACACAC
ACCGTACTCTGTATAAATGAAGCATCACACACACACACACTCACACACACACACACACACACACACACAA
ACACACACACACACACACACACACACACACACACACACACACACACACACACACCATACTCTGTATAAAT
GAAGCATCACACACACACACACTCACACACACACACACACACACACACACACACACACACACACACACAC
ACACACCATACTCTGTATAAATGAAGCATCACACACACACACACACATACTCCATACTCTGAATTAATGA
AGTGTAATGTTTCCTCTACCAGATGTTTCCGAACTTCTCCAAGGACTCCACCAGATCAGCCTCCACCACA
GACTCACAGAGACCCCGAACATGGACCACAGGAGACGGAGCGATTCTGTGTGTCTCTGAGCTGCTGTCCT
GAGGAGGAAGACATGACAGATCAACACTACACTCACTCATTCATTCATTCATTCATTCATTCATTCATTC
ATTCATTCATTCATTCATTCATTCATTCACTCATTCATTCATTCATTGACAGAGCAATTCTGTGCATCTC
TGAGCAGCTGTCCTGAAGAGACATGACAGATCAACACCACATTCATTCATTCATTCATTCATTCATTCAT
TCACTCATTCATTCATTCATTGACAGAGCAATTCTGTGCATCTCTAAGCAGCTGTCTTGAAGAGACATGA
CAGATCAACACCACATTCATTTATTAATTAATTAATTCATTCATTCATTCCTTCATTTATCAGTTCCTTC
AGTCATTCATTAATTCAGTTATTAAATCATCAACAGGGCTATTTTGTGTGTCTCTGAGCGCTATCCTGAG
GACGGA