ZFLNCG07386
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR592701 | pineal gland | normal | 9.49 |
| ERR023144 | brain | normal | 8.66 |
| SRR592699 | pineal gland | normal | 7.19 |
| SRR592700 | pineal gland | normal | 5.73 |
| SRR592702 | pineal gland | normal | 4.35 |
| SRR519732 | 7 dpf | FETOH treatment | 3.49 |
| SRR519727 | 6 dpf | FETOH treatment | 3.47 |
| SRR519752 | 7 dpf | VD3 treatment | 3.40 |
| SRR519722 | 4 dpf | FETOH treatment | 3.05 |
| SRR519747 | 6 dpf | VD3 treatment | 2.84 |
Express in tissues
Correlated coding gene
Download
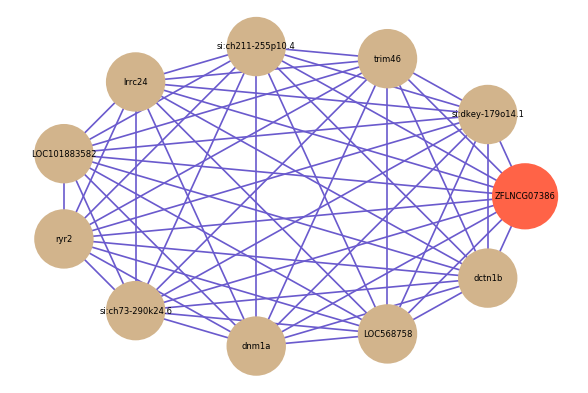
| gene | correlation coefficent |
|---|---|
| si:dkey-179o14.1 | 0.72 |
| trim46 | 0.71 |
| si:ch211-255p10.4 | 0.71 |
| lrrc24 | 0.71 |
| LOC101883582 | 0.71 |
| ryr2 | 0.71 |
| si:ch73-290k24.6 | 0.71 |
| dnm1a | 0.71 |
| LOC568758 | 0.70 |
| dctn1b | 0.70 |
Gene Ontology
Download
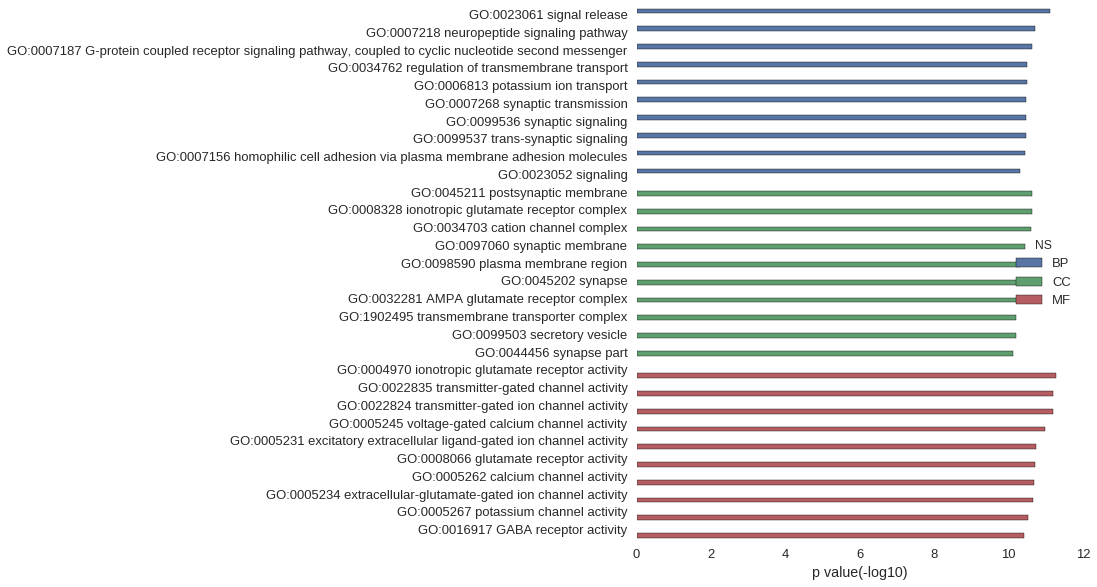
| GO | P value |
|---|---|
| GO:0023061 | 7.62e-12 |
| GO:0007218 | 2.00e-11 |
| GO:0007187 | 2.41e-11 |
| GO:0034762 | 3.27e-11 |
| GO:0006813 | 3.27e-11 |
| GO:0007268 | 3.36e-11 |
| GO:0099536 | 3.36e-11 |
| GO:0099537 | 3.36e-11 |
| GO:0007156 | 3.62e-11 |
| GO:0023052 | 4.91e-11 |
| GO:0045211 | 2.30e-11 |
| GO:0008328 | 2.35e-11 |
| GO:0034703 | 2.45e-11 |
| GO:0097060 | 3.69e-11 |
| GO:0098590 | 5.04e-11 |
| GO:0045202 | 5.34e-11 |
| GO:0032281 | 5.78e-11 |
| GO:1902495 | 6.24e-11 |
| GO:0099503 | 6.46e-11 |
| GO:0044456 | 7.58e-11 |
| GO:0004970 | 5.34e-12 |
| GO:0022835 | 6.21e-12 |
| GO:0022824 | 6.21e-12 |
| GO:0005245 | 1.03e-11 |
| GO:0005231 | 1.80e-11 |
| GO:0008066 | 1.90e-11 |
| GO:0005262 | 2.09e-11 |
| GO:0005234 | 2.26e-11 |
| GO:0005267 | 2.91e-11 |
| GO:0016917 | 3.85e-11 |
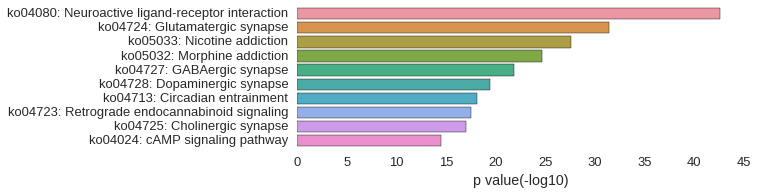
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG07386
TAAAAATACTTGTGTGCTGTTAGAGAGTTGCTGGGATATGAGAAAGTTCACTTGATGAAGAATGACTGTG
ACCTTCAGCTGCAGTGACTCTGGAAACTCCTACCTTTGAGTTTGATGTGGACTCCAGAAAACACAGTCTG
TTCCCGAAGCCCTCGGCGGCCAGACAGAGCTTCTGCTGCTCCTTATGGAGCGTCGCCGAGCACTGGAGGA
CCACCTCATCATCCTGAACACACAAATAAAACACAGATCACAATCTTCATCATCATCATCATCACTGGAG
GACCACCTCATCATCCTGAACACACAAATAAAACACAGATCACAATCTTCATCATCATCATCATCATCAC
TGGAGGACCATCTCATCATCCTGAACACACAAATAAAACACAGATCACAATCTTCATCATCATCATCATC
ACTGGAGGACCACCTCATCATCCTGAACACACACAAAACACACACACACACAAAACACACATAATCATCA
TCATCATCACCTTCATTATCACTGGAGGACCACCTCATCATCCTGAACACACACACACACACACACACAC
ACACACACACACACACATCTTCATCATCAACACTGGAGGACCACCTCATCATCCTGAACACACAAATAAA
ACACAGATCACAATCTTCATCATCATCATCATCACTGGAGGACCACCTCATCATCCTGAACACACACAAA
AAACACACACACACACACACACACACACACACAAAAGACACATAATCATCATCATCATCATCACCTTCAT
TATCACTGGAGGACCACCTCATCATCCTGAACACACACACACACACACACACACACATCTTCATCATCAA
CACTGGAGGACCACCTCATCATCCT
TAAAAATACTTGTGTGCTGTTAGAGAGTTGCTGGGATATGAGAAAGTTCACTTGATGAAGAATGACTGTG
ACCTTCAGCTGCAGTGACTCTGGAAACTCCTACCTTTGAGTTTGATGTGGACTCCAGAAAACACAGTCTG
TTCCCGAAGCCCTCGGCGGCCAGACAGAGCTTCTGCTGCTCCTTATGGAGCGTCGCCGAGCACTGGAGGA
CCACCTCATCATCCTGAACACACAAATAAAACACAGATCACAATCTTCATCATCATCATCATCACTGGAG
GACCACCTCATCATCCTGAACACACAAATAAAACACAGATCACAATCTTCATCATCATCATCATCATCAC
TGGAGGACCATCTCATCATCCTGAACACACAAATAAAACACAGATCACAATCTTCATCATCATCATCATC
ACTGGAGGACCACCTCATCATCCTGAACACACACAAAACACACACACACACAAAACACACATAATCATCA
TCATCATCACCTTCATTATCACTGGAGGACCACCTCATCATCCTGAACACACACACACACACACACACAC
ACACACACACACACACATCTTCATCATCAACACTGGAGGACCACCTCATCATCCTGAACACACAAATAAA
ACACAGATCACAATCTTCATCATCATCATCATCACTGGAGGACCACCTCATCATCCTGAACACACACAAA
AAACACACACACACACACACACACACACACACAAAAGACACATAATCATCATCATCATCATCACCTTCAT
TATCACTGGAGGACCACCTCATCATCCTGAACACACACACACACACACACACACACATCTTCATCATCAA
CACTGGAGGACCACCTCATCATCCT