ZFLNCG07529
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR592703 | pineal gland | normal | 83.07 |
| SRR1648855 | brain | normal | 16.25 |
| SRR1534351 | larvae | 500 ug/l BDE47 treatment | 11.25 |
| SRR1028003 | head | normal | 9.96 |
| SRR1028002 | head | normal | 9.80 |
| ERR594417 | retina | normal | 7.44 |
| SRR1565811 | brain | normal | 6.83 |
| SRR726541 | 5 dpf | infection with Mycobacterium marinum | 6.83 |
| SRR1565805 | brain | normal | 6.34 |
| SRR1028004 | head | normal | 5.99 |
Express in tissues
Correlated coding gene
Download
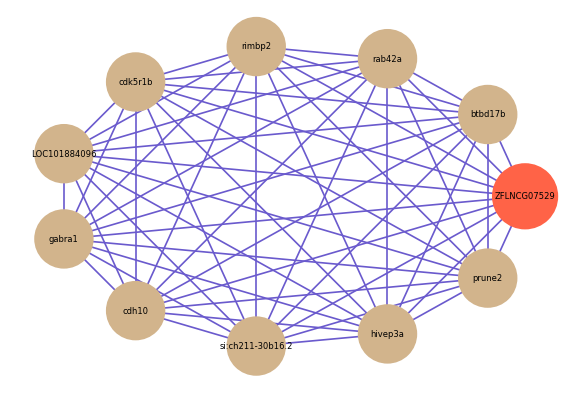
| gene | correlation coefficent |
|---|---|
| btbd17b | 0.75 |
| rab42a | 0.75 |
| rimbp2 | 0.75 |
| cdk5r1b | 0.75 |
| LOC101884096 | 0.75 |
| gabra1 | 0.75 |
| cdh10 | 0.74 |
| si:ch211-30b16.2 | 0.74 |
| hivep3a | 0.74 |
| prune2 | 0.74 |
Gene Ontology
Download
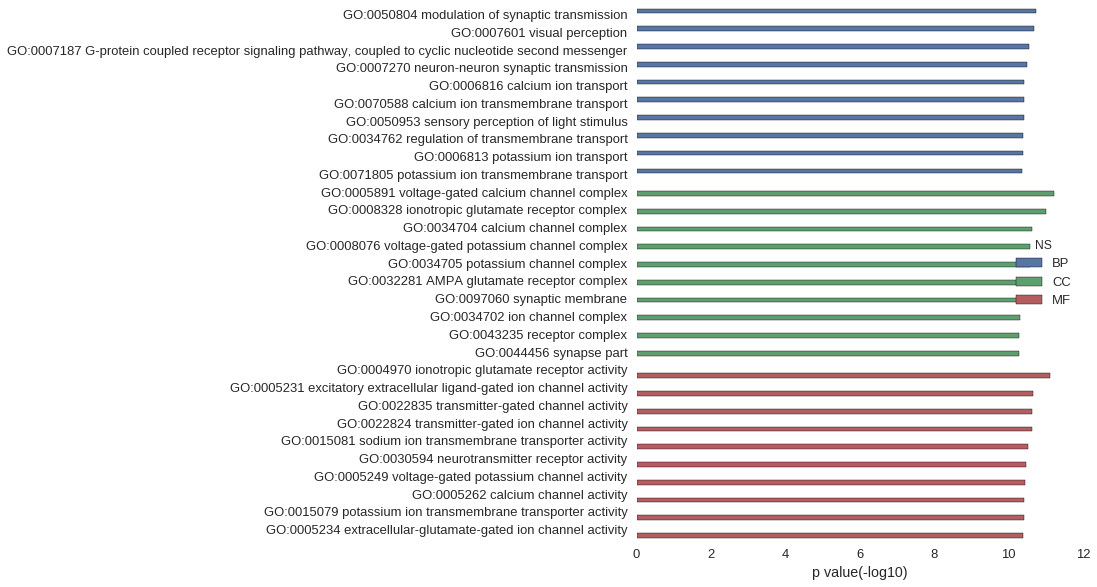
| GO | P value |
|---|---|
| GO:0050804 | 1.82e-11 |
| GO:0007601 | 2.10e-11 |
| GO:0007187 | 2.89e-11 |
| GO:0007270 | 3.22e-11 |
| GO:0006816 | 3.78e-11 |
| GO:0070588 | 3.80e-11 |
| GO:0050953 | 3.80e-11 |
| GO:0034762 | 4.05e-11 |
| GO:0006813 | 4.05e-11 |
| GO:0071805 | 4.22e-11 |
| GO:0005891 | 5.87e-12 |
| GO:0008328 | 1.00e-11 |
| GO:0034704 | 2.32e-11 |
| GO:0008076 | 2.62e-11 |
| GO:0034705 | 2.62e-11 |
| GO:0032281 | 3.81e-11 |
| GO:0097060 | 4.70e-11 |
| GO:0034702 | 5.05e-11 |
| GO:0043235 | 5.10e-11 |
| GO:0044456 | 5.15e-11 |
| GO:0004970 | 7.47e-12 |
| GO:0005231 | 2.26e-11 |
| GO:0022835 | 2.32e-11 |
| GO:0022824 | 2.32e-11 |
| GO:0015081 | 3.09e-11 |
| GO:0030594 | 3.37e-11 |
| GO:0005249 | 3.51e-11 |
| GO:0005262 | 3.80e-11 |
| GO:0015079 | 3.83e-11 |
| GO:0005234 | 4.06e-11 |
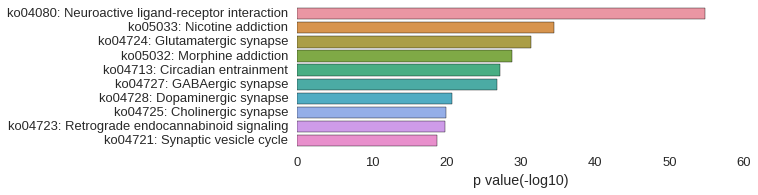
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG07529
TCAGATGAGCAGCTCTGGATATACACAAGGATTGTGTGTCTTGCCGATCAGGGGACGCCTGTGGATCATG
TTTCCGTGAATACTGACAGAAATAAAGCGTATTTCACGTATCTGCTCTCTCACCGTGTCAGTCTGTATGC
ATTTGCGCGCGCACTGGGAGTTTCCGTCAATCTACGGATGAGACACCGCTGGAAGTCAGAGCTTCTGTGG
CACAACTTTTCTGATTTATTCGTCTCAGAGGATCGGCAATTTTGTAAAACG
TCAGATGAGCAGCTCTGGATATACACAAGGATTGTGTGTCTTGCCGATCAGGGGACGCCTGTGGATCATG
TTTCCGTGAATACTGACAGAAATAAAGCGTATTTCACGTATCTGCTCTCTCACCGTGTCAGTCTGTATGC
ATTTGCGCGCGCACTGGGAGTTTCCGTCAATCTACGGATGAGACACCGCTGGAAGTCAGAGCTTCTGTGG
CACAACTTTTCTGATTTATTCGTCTCAGAGGATCGGCAATTTTGTAAAACG