ZFLNCG07588
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR592702 | pineal gland | normal | 55.42 |
| SRR1648856 | brain | normal | 51.20 |
| ERR023144 | brain | normal | 49.68 |
| SRR592701 | pineal gland | normal | 33.48 |
| SRR592699 | pineal gland | normal | 33.43 |
| SRR1562529 | intestine and pancreas | normal | 32.13 |
| ERR023146 | head kidney | normal | 31.46 |
| SRR592703 | pineal gland | normal | 28.14 |
| SRR372802 | 5 dpf | normal | 24.84 |
| SRR1039571 | gastrointestinal | diet 0.1 NPM | 24.82 |
Express in tissues
Correlated coding gene
Download
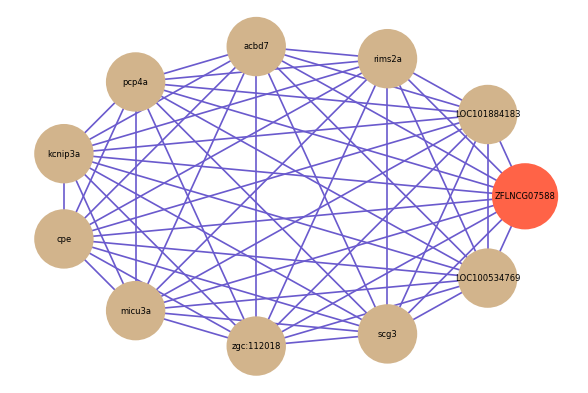
| gene | correlation coefficent |
|---|---|
| LOC101884183 | 0.84 |
| rims2a | 0.79 |
| acbd7 | 0.79 |
| pcp4a | 0.77 |
| kcnip3a | 0.77 |
| cpe | 0.77 |
| micu3a | 0.77 |
| zgc:112018 | 0.77 |
| scg3 | 0.77 |
| LOC100534769 | 0.76 |
Gene Ontology
Download
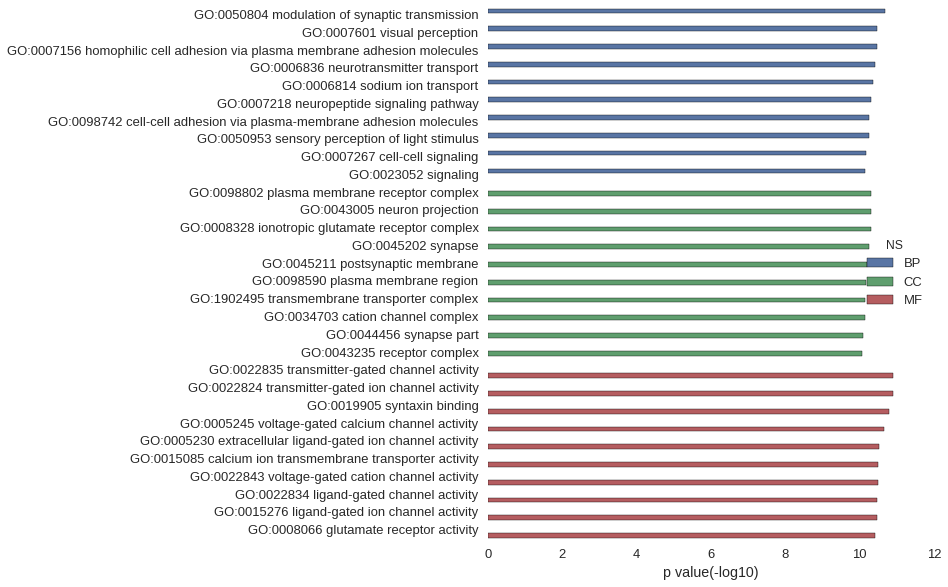
| GO | P value |
|---|---|
| GO:0050804 | 2.07e-11 |
| GO:0007601 | 3.44e-11 |
| GO:0007156 | 3.51e-11 |
| GO:0006836 | 3.80e-11 |
| GO:0006814 | 4.43e-11 |
| GO:0007218 | 5.06e-11 |
| GO:0098742 | 5.47e-11 |
| GO:0050953 | 5.79e-11 |
| GO:0007267 | 6.64e-11 |
| GO:0023052 | 7.23e-11 |
| GO:0098802 | 4.84e-11 |
| GO:0043005 | 4.84e-11 |
| GO:0008328 | 4.92e-11 |
| GO:0045202 | 5.47e-11 |
| GO:0045211 | 6.54e-11 |
| GO:0098590 | 6.89e-11 |
| GO:1902495 | 7.30e-11 |
| GO:0034703 | 7.37e-11 |
| GO:0044456 | 8.18e-11 |
| GO:0043235 | 8.85e-11 |
| GO:0022835 | 1.27e-11 |
| GO:0022824 | 1.27e-11 |
| GO:0019905 | 1.58e-11 |
| GO:0005245 | 2.24e-11 |
| GO:0005230 | 2.97e-11 |
| GO:0015085 | 3.13e-11 |
| GO:0022843 | 3.16e-11 |
| GO:0022834 | 3.44e-11 |
| GO:0015276 | 3.44e-11 |
| GO:0008066 | 3.85e-11 |
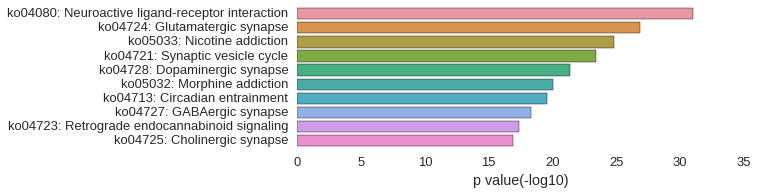
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG07588
ATCTGTCATATTGCTTTTATTGTTACATTAGAATACAGACAAAAACAATGCTGCTGGTGAAACATTTGAT
TAGCTTTTGACAAATAACTAAACAAACTGGACTCTACTTCTGTATCAACATGTGTAAATCCAAATATAAT
CCTGCTTCTATTTCAGAAATCCGTACTTGTTACACCATGCTCCAGTAGACCCTTTATAAATTATTTTTAA
AAGGTGAATCTGACTGGAAATTGTATCATTCAGAAATGTAATGTCTAATCAACAGTAAGAACTAATAAGC
TCACCTGCTACATCTAATATGCTAATATTTATTAGTACTGTTATATTAGTATAATATTTA
ATCTGTCATATTGCTTTTATTGTTACATTAGAATACAGACAAAAACAATGCTGCTGGTGAAACATTTGAT
TAGCTTTTGACAAATAACTAAACAAACTGGACTCTACTTCTGTATCAACATGTGTAAATCCAAATATAAT
CCTGCTTCTATTTCAGAAATCCGTACTTGTTACACCATGCTCCAGTAGACCCTTTATAAATTATTTTTAA
AAGGTGAATCTGACTGGAAATTGTATCATTCAGAAATGTAATGTCTAATCAACAGTAAGAACTAATAAGC
TCACCTGCTACATCTAATATGCTAATATTTATTAGTACTGTTATATTAGTATAATATTTA