ZFLNCG07663
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1048061 | pineal gland | dark | 3325.18 |
| SRR1048059 | pineal gland | light | 3205.06 |
| SRR592702 | pineal gland | normal | 2174.55 |
| SRR592701 | pineal gland | normal | 1949.40 |
| SRR592703 | pineal gland | normal | 1885.52 |
| SRR1028002 | head | normal | 1820.59 |
| SRR1028003 | head | normal | 1674.85 |
| SRR592700 | pineal gland | normal | 1396.08 |
| SRR1028004 | head | normal | 1255.22 |
| SRR592699 | pineal gland | normal | 912.81 |
Express in tissues
Correlated coding gene
Download
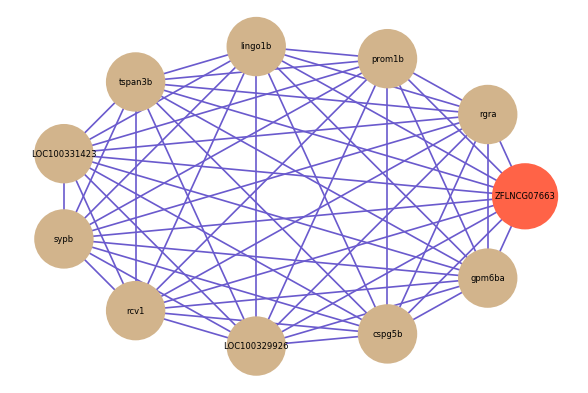
| gene | correlation coefficent |
|---|---|
| rgra | 0.89 |
| prom1b | 0.87 |
| lingo1b | 0.86 |
| tspan3b | 0.85 |
| LOC100331423 | 0.85 |
| sypb | 0.85 |
| rcv1 | 0.84 |
| LOC100329926 | 0.84 |
| cspg5b | 0.84 |
| gpm6ba | 0.84 |
Gene Ontology
Download
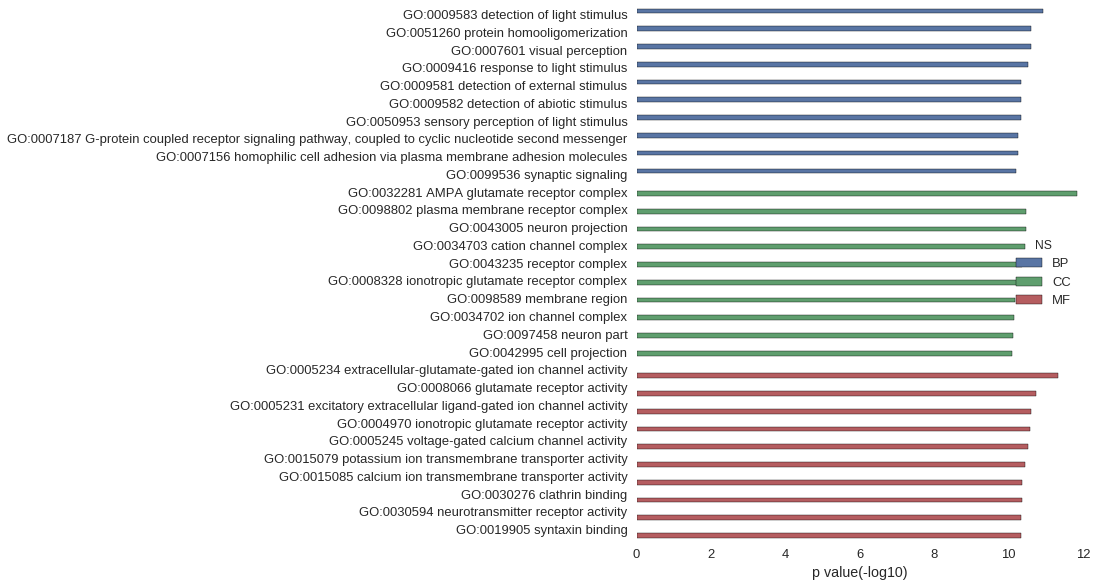
| GO | P value |
|---|---|
| GO:0009583 | 1.18e-11 |
| GO:0051260 | 2.45e-11 |
| GO:0007601 | 2.55e-11 |
| GO:0009416 | 2.97e-11 |
| GO:0009581 | 4.59e-11 |
| GO:0009582 | 4.59e-11 |
| GO:0050953 | 4.72e-11 |
| GO:0007187 | 5.56e-11 |
| GO:0007156 | 5.59e-11 |
| GO:0099536 | 6.49e-11 |
| GO:0032281 | 1.46e-12 |
| GO:0098802 | 3.34e-11 |
| GO:0043005 | 3.34e-11 |
| GO:0034703 | 3.70e-11 |
| GO:0043235 | 4.36e-11 |
| GO:0008328 | 4.48e-11 |
| GO:0098589 | 6.88e-11 |
| GO:0034702 | 7.03e-11 |
| GO:0097458 | 7.49e-11 |
| GO:0042995 | 7.96e-11 |
| GO:0005234 | 4.70e-12 |
| GO:0008066 | 1.85e-11 |
| GO:0005231 | 2.49e-11 |
| GO:0004970 | 2.60e-11 |
| GO:0005245 | 2.95e-11 |
| GO:0015079 | 3.53e-11 |
| GO:0015085 | 4.24e-11 |
| GO:0030276 | 4.44e-11 |
| GO:0030594 | 4.58e-11 |
| GO:0019905 | 4.59e-11 |
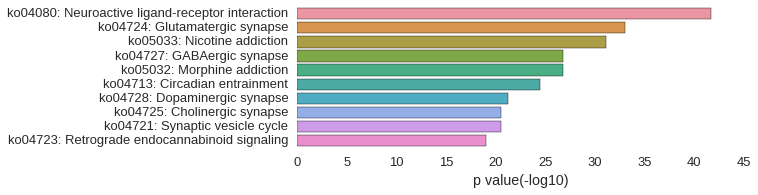
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG07663
CAGCCAGCAATGTTTATTTCACTGTTGTGCATCATGGCTAAAATATACAAATCAGTCGCTTCACAAGACA
TCTGTTAAACATTCACATCATTCACAGGTTAATTCAAGTGCACTTGAGGAAGACAAGCTTAAAGTGGACA
CAAAGACTAGTCCGTTTCTAGAATGACAAGTTAAACGAAACCACAGAAAGTGTCAGTAACTAGA
CAGCCAGCAATGTTTATTTCACTGTTGTGCATCATGGCTAAAATATACAAATCAGTCGCTTCACAAGACA
TCTGTTAAACATTCACATCATTCACAGGTTAATTCAAGTGCACTTGAGGAAGACAAGCTTAAAGTGGACA
CAAAGACTAGTCCGTTTCTAGAATGACAAGTTAAACGAAACCACAGAAAGTGTCAGTAACTAGA