ZFLNCG07961
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR592702 | pineal gland | normal | 78.14 |
| SRR592703 | pineal gland | normal | 73.13 |
| SRR592701 | pineal gland | normal | 56.88 |
| SRR1048061 | pineal gland | dark | 23.54 |
| SRR1569495 | embryo | normal | 22.00 |
| ERR023144 | brain | normal | 19.62 |
| SRR592699 | pineal gland | normal | 17.51 |
| SRR1048059 | pineal gland | light | 14.44 |
| SRR065196 | 3 dpf | normal | 12.42 |
| SRR1565809 | brain | normal | 11.82 |
Express in tissues
Correlated coding gene
Download
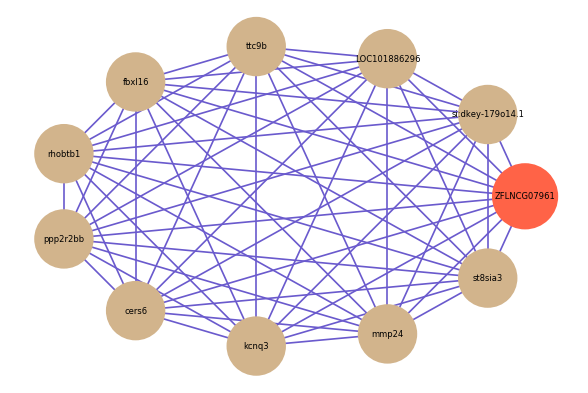
| gene | correlation coefficent |
|---|---|
| si:dkey-179o14.1 | 0.81 |
| LOC101886296 | 0.81 |
| ttc9b | 0.81 |
| fbxl16 | 0.80 |
| rhobtb1 | 0.80 |
| ppp2r2bb | 0.80 |
| cers6 | 0.79 |
| kcnq3 | 0.79 |
| mmp24 | 0.79 |
| st8sia3 | 0.79 |
Gene Ontology
Download
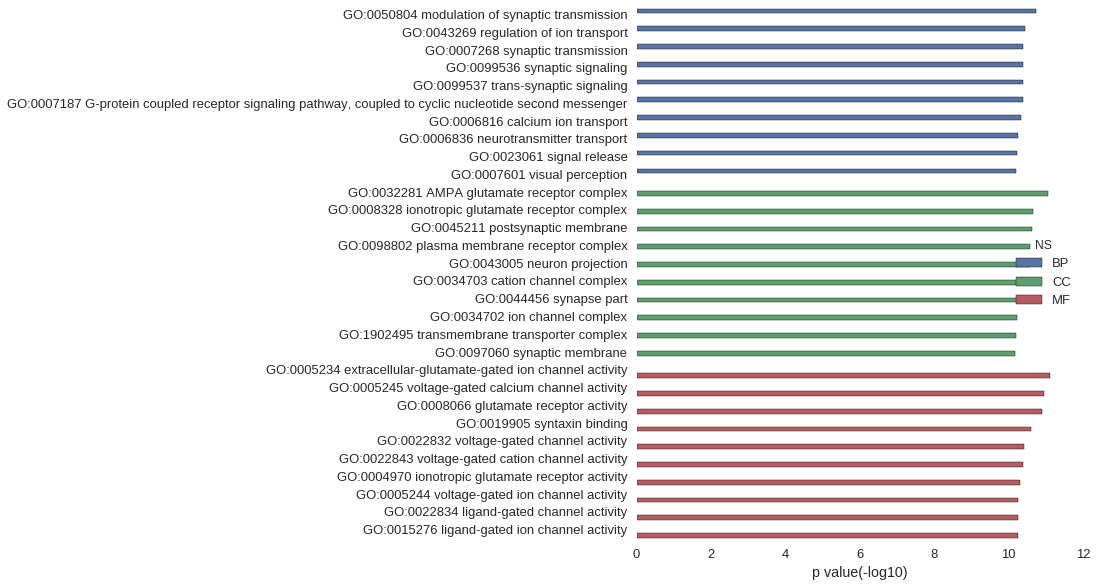
| GO | P value |
|---|---|
| GO:0050804 | 1.83e-11 |
| GO:0043269 | 3.65e-11 |
| GO:0007268 | 4.07e-11 |
| GO:0099536 | 4.07e-11 |
| GO:0099537 | 4.07e-11 |
| GO:0007187 | 4.17e-11 |
| GO:0006816 | 4.72e-11 |
| GO:0006836 | 5.45e-11 |
| GO:0023061 | 6.03e-11 |
| GO:0007601 | 6.36e-11 |
| GO:0032281 | 8.61e-12 |
| GO:0008328 | 2.14e-11 |
| GO:0045211 | 2.27e-11 |
| GO:0098802 | 2.62e-11 |
| GO:0043005 | 2.62e-11 |
| GO:0034703 | 3.55e-11 |
| GO:0044456 | 5.58e-11 |
| GO:0034702 | 6.08e-11 |
| GO:1902495 | 6.20e-11 |
| GO:0097060 | 6.55e-11 |
| GO:0005234 | 7.66e-12 |
| GO:0005245 | 1.13e-11 |
| GO:0008066 | 1.27e-11 |
| GO:0019905 | 2.49e-11 |
| GO:0022832 | 3.75e-11 |
| GO:0022843 | 4.12e-11 |
| GO:0004970 | 4.81e-11 |
| GO:0005244 | 5.58e-11 |
| GO:0022834 | 5.63e-11 |
| GO:0015276 | 5.63e-11 |
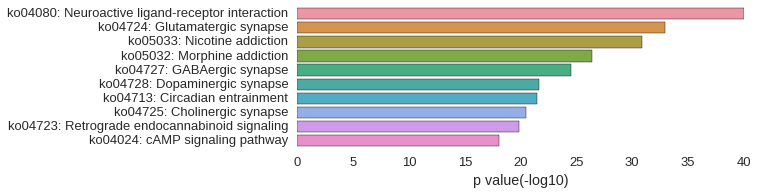
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG07961
AGCCGGAGAGAGACGCGCGAGGGGAACAGCGAGCGCGCTTACGAGACTGCGCCCATCACACCGAGATCCA
GCAGCCCGAACAATGGCAGCATCCAGCACCTGATTTACAGATGGAGACGTGCTGGAGCTAAAGAGCGCCG
CTTTTTAACGACACGGCCCTGCTGTCATCGGAGATGGAGCGATAGAGGAAAGAGAATCAGCGGGCTGTTC
TCCATCAAGTGTGCCGCAAGCCTGCGTTTGAGTGAAGGTCTCGGCCGTTAGATCGCCCCCTGGGGGCTGG
CTGCAGTACAAGTCATAAAGCCCGCCTCCTC
AGCCGGAGAGAGACGCGCGAGGGGAACAGCGAGCGCGCTTACGAGACTGCGCCCATCACACCGAGATCCA
GCAGCCCGAACAATGGCAGCATCCAGCACCTGATTTACAGATGGAGACGTGCTGGAGCTAAAGAGCGCCG
CTTTTTAACGACACGGCCCTGCTGTCATCGGAGATGGAGCGATAGAGGAAAGAGAATCAGCGGGCTGTTC
TCCATCAAGTGTGCCGCAAGCCTGCGTTTGAGTGAAGGTCTCGGCCGTTAGATCGCCCCCTGGGGGCTGG
CTGCAGTACAAGTCATAAAGCCCGCCTCCTC