ZFLNCG08289
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR594769 | blood | normal | 133.76 |
| SRR658545 | 6 somite | Gata6 morphant | 127.87 |
| SRR658541 | 6 somite | normal | 126.04 |
| SRR658543 | 6 somite | Gata5 morphan | 103.46 |
| SRR658533 | bud | normal | 95.46 |
| SRR891511 | brain | normal | 91.89 |
| SRR594771 | endothelium | normal | 90.58 |
| SRR658547 | 6 somite | Gata5/6 morphant | 79.85 |
| SRR1035978 | 13 hpf | rx3-/- | 79.17 |
| SRR658539 | bud | Gata5/6 morphant | 77.71 |
Express in tissues
Correlated coding gene
Download
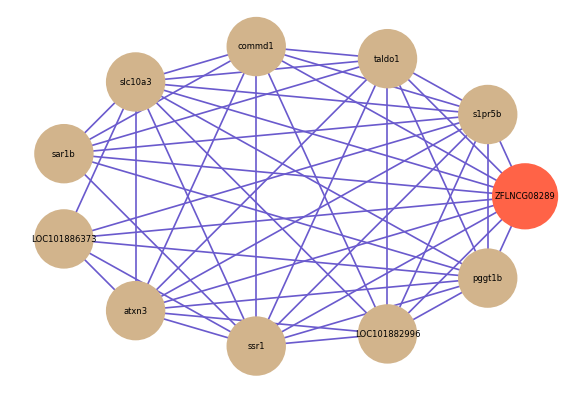
| gene | correlation coefficent |
|---|---|
| s1pr5b | 0.66 |
| taldo1 | 0.62 |
| commd1 | 0.62 |
| slc10a3 | 0.61 |
| sar1b | 0.59 |
| LOC101886373 | 0.58 |
| atxn3 | 0.58 |
| ssr1 | 0.57 |
| LOC101882996 | 0.56 |
| pggt1b | 0.56 |
Gene Ontology
Download
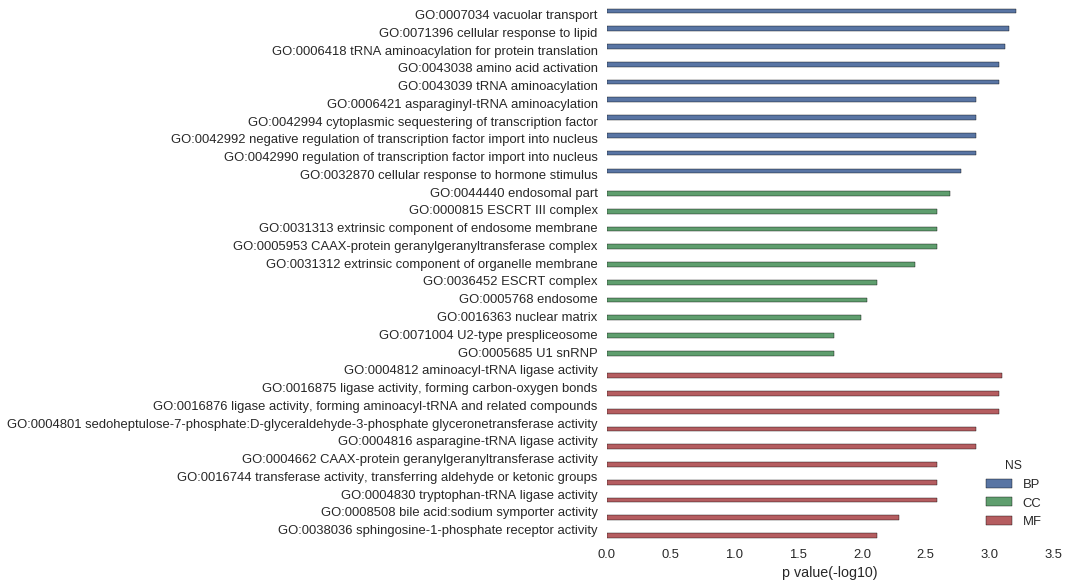
| GO | P value |
|---|---|
| GO:0007034 | 6.15e-04 |
| GO:0071396 | 7.03e-04 |
| GO:0006418 | 7.49e-04 |
| GO:0043038 | 8.46e-04 |
| GO:0043039 | 8.46e-04 |
| GO:0006421 | 1.28e-03 |
| GO:0042994 | 1.28e-03 |
| GO:0042992 | 1.28e-03 |
| GO:0042990 | 1.28e-03 |
| GO:0032870 | 1.68e-03 |
| GO:0044440 | 2.05e-03 |
| GO:0000815 | 2.56e-03 |
| GO:0031313 | 2.56e-03 |
| GO:0005953 | 2.56e-03 |
| GO:0031312 | 3.83e-03 |
| GO:0036452 | 7.65e-03 |
| GO:0005768 | 9.15e-03 |
| GO:0016363 | 1.02e-02 |
| GO:0071004 | 1.65e-02 |
| GO:0005685 | 1.65e-02 |
| GO:0004812 | 7.97e-04 |
| GO:0016875 | 8.46e-04 |
| GO:0016876 | 8.46e-04 |
| GO:0004801 | 1.28e-03 |
| GO:0004816 | 1.28e-03 |
| GO:0004662 | 2.56e-03 |
| GO:0016744 | 2.56e-03 |
| GO:0004830 | 2.56e-03 |
| GO:0008508 | 5.11e-03 |
| GO:0038036 | 7.65e-03 |
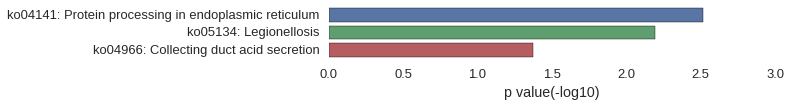
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG08289
TGTGTGTGTGTGTGTGTGTGTAGCGCGTCTGTAAGGAGGTGTACCAGCTGAAGGGAGGTGTTCATAAGTA
CCTGGAGCAGTTCCCTGACGGATTCTACCGCGGTAAACTCTTCGTGTTCGACGAGCGCTACGCCATCGCC
TACAACCAGGACGTCATCTCAGGTGTGTGTTCTGTCCAGTACAGGTGTGTGTTGGTCATGCAGCACAGGT
GTGTGTGTGTGTGATGTCTCTCTCTCTCTCTCCGCAGTGTGTCGGTACTGCTCTCAGCCGTGGGATCAGT
ACGTGCGCTGCTCTACAGGTGTGTGTGGTCAGCTGCTGCTGTCGTGTGTGTCCTGCCGTGTGTGGTCTGA
CCGCCTGCTGTGAAGAGTGTCAGGAGTCCAGCCGAGAGCAGTGCCAGTGCACCCGGAGCCGGCCGCGCAT
ACCCACACACACACACACACACACTGCTCAGAACAGTCAATAAATCACACACACACACACACACACTGCT
CAGAACAGTCAGTAGATCTCACACACACACACACACACACGCACACACACACACACTGCTCAGAACAGTC
AGTAGATCAACAATCATGACACACACACACACACACACACACACACACACACACACACTGCTCAGAACAC
TCAAATCAACAATCATGATACACACACACTCAATAAATGTGCAGTCATGATCCTCCGTCTGTTTATTTCT
CAATTAAAGCACACATTTACATATAAATCAACACTTTGATC
TGTGTGTGTGTGTGTGTGTGTAGCGCGTCTGTAAGGAGGTGTACCAGCTGAAGGGAGGTGTTCATAAGTA
CCTGGAGCAGTTCCCTGACGGATTCTACCGCGGTAAACTCTTCGTGTTCGACGAGCGCTACGCCATCGCC
TACAACCAGGACGTCATCTCAGGTGTGTGTTCTGTCCAGTACAGGTGTGTGTTGGTCATGCAGCACAGGT
GTGTGTGTGTGTGATGTCTCTCTCTCTCTCTCCGCAGTGTGTCGGTACTGCTCTCAGCCGTGGGATCAGT
ACGTGCGCTGCTCTACAGGTGTGTGTGGTCAGCTGCTGCTGTCGTGTGTGTCCTGCCGTGTGTGGTCTGA
CCGCCTGCTGTGAAGAGTGTCAGGAGTCCAGCCGAGAGCAGTGCCAGTGCACCCGGAGCCGGCCGCGCAT
ACCCACACACACACACACACACACTGCTCAGAACAGTCAATAAATCACACACACACACACACACACTGCT
CAGAACAGTCAGTAGATCTCACACACACACACACACACACGCACACACACACACACTGCTCAGAACAGTC
AGTAGATCAACAATCATGACACACACACACACACACACACACACACACACACACACACTGCTCAGAACAC
TCAAATCAACAATCATGATACACACACACTCAATAAATGTGCAGTCATGATCCTCCGTCTGTTTATTTCT
CAATTAAAGCACACATTTACATATAAATCAACACTTTGATC