ZFLNCG08351
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1647684 | spleen | SVCV treatment | 434.38 |
| SRR1647683 | spleen | SVCV treatment | 164.67 |
| SRR957180 | heart | 7 days after heart tip amputation | 155.13 |
| ERR023146 | head kidney | normal | 98.94 |
| SRR1647681 | head kidney | SVCV treatment | 91.85 |
| SRR1647682 | spleen | normal | 77.69 |
| ERR023145 | heart | normal | 50.84 |
| SRR1647680 | head kidney | SVCV treatment | 38.77 |
| ERR023143 | swim bladder | normal | 36.60 |
| SRR700534 | heart | control morpholino | 29.32 |
Express in tissues
Correlated coding gene
Download
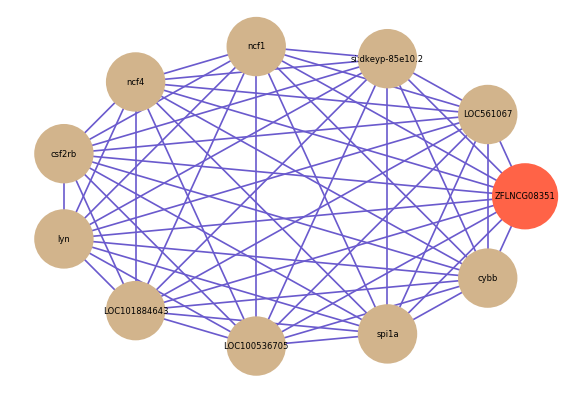
| gene | correlation coefficent |
|---|---|
| LOC561067 | 0.73 |
| si:dkeyp-85e10.2 | 0.65 |
| ncf1 | 0.64 |
| ncf4 | 0.64 |
| csf2rb | 0.64 |
| lyn | 0.63 |
| LOC101884643 | 0.63 |
| LOC100536705 | 0.63 |
| spi1a | 0.63 |
| cybb | 0.62 |
Gene Ontology
Download
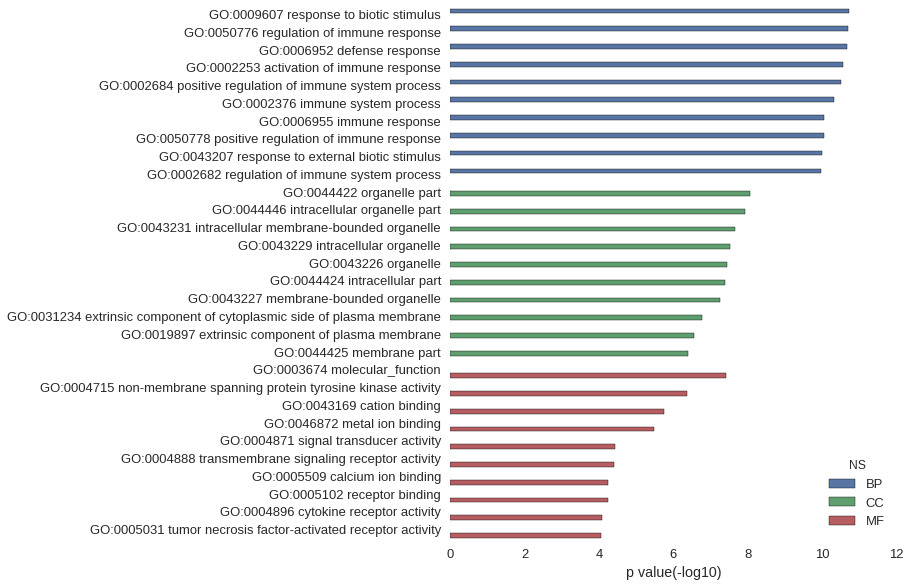
| GO | P value |
|---|---|
| GO:0009607 | 1.90e-11 |
| GO:0050776 | 2.03e-11 |
| GO:0006952 | 2.14e-11 |
| GO:0002253 | 2.83e-11 |
| GO:0002684 | 3.15e-11 |
| GO:0002376 | 4.93e-11 |
| GO:0006955 | 8.69e-11 |
| GO:0050778 | 8.72e-11 |
| GO:0043207 | 9.90e-11 |
| GO:0002682 | 1.06e-10 |
| GO:0044422 | 8.72e-09 |
| GO:0044446 | 1.22e-08 |
| GO:0043231 | 2.22e-08 |
| GO:0043229 | 3.00e-08 |
| GO:0043226 | 3.54e-08 |
| GO:0044424 | 4.13e-08 |
| GO:0043227 | 5.45e-08 |
| GO:0031234 | 1.70e-07 |
| GO:0019897 | 2.78e-07 |
| GO:0044425 | 4.03e-07 |
| GO:0003674 | 3.79e-08 |
| GO:0004715 | 4.41e-07 |
| GO:0043169 | 1.76e-06 |
| GO:0046872 | 3.34e-06 |
| GO:0004871 | 3.61e-05 |
| GO:0004888 | 3.99e-05 |
| GO:0005509 | 5.85e-05 |
| GO:0005102 | 5.87e-05 |
| GO:0004896 | 8.44e-05 |
| GO:0005031 | 8.75e-05 |
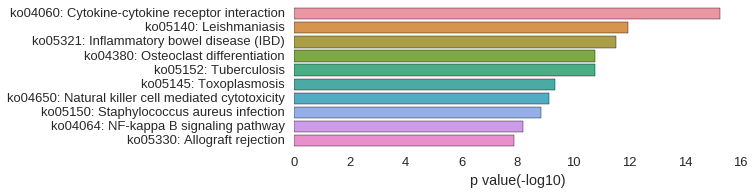
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG08351
TTTTGTGTTAGTTAACATACTTACAATAACTTGTAACTAGCGATATGGGTAGGGTTAGCAAGAGTCTTAG
AGTCCTGGTCTCGTAGCAGAACCTTTGGCGTATGATTTTGGTATTTAAACTGCCTCTTTGCACAATAATC
TAGATTAGTTTAGTTAAGTTTATTTCATATATTTGTACAGCAATGATTTACAACATGACATGCTTTGCCA
GATTTAGCTAGAAAGCTAATTAGCATCTGTAGACCCTGGGCAGGATTTTGATTGTTACAAAATTAACCAC
A
TTTTGTGTTAGTTAACATACTTACAATAACTTGTAACTAGCGATATGGGTAGGGTTAGCAAGAGTCTTAG
AGTCCTGGTCTCGTAGCAGAACCTTTGGCGTATGATTTTGGTATTTAAACTGCCTCTTTGCACAATAATC
TAGATTAGTTTAGTTAAGTTTATTTCATATATTTGTACAGCAATGATTTACAACATGACATGCTTTGCCA
GATTTAGCTAGAAAGCTAATTAGCATCTGTAGACCCTGGGCAGGATTTTGATTGTTACAAAATTAACCAC
A