ZFLNCG08586
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR700534 | heart | control morpholino | 96.79 |
| ERR023143 | swim bladder | normal | 43.13 |
| ERR023146 | head kidney | normal | 36.92 |
| SRR1647680 | head kidney | SVCV treatment | 36.65 |
| SRR1647684 | spleen | SVCV treatment | 35.98 |
| SRR1647683 | spleen | SVCV treatment | 35.43 |
| ERR023145 | heart | normal | 32.66 |
| SRR1647679 | head kidney | normal | 27.08 |
| SRR700537 | heart | Gata4 morpholino | 25.86 |
| SRR1647681 | head kidney | SVCV treatment | 22.20 |
Express in tissues
Correlated coding gene
Download
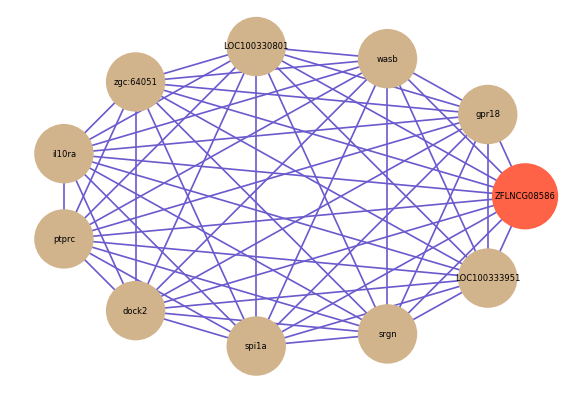
| gene | correlation coefficent |
|---|---|
| gpr18 | 0.75 |
| wasb | 0.74 |
| LOC100330801 | 0.74 |
| zgc:64051 | 0.74 |
| il10ra | 0.74 |
| ptprc | 0.73 |
| dock2 | 0.73 |
| spi1a | 0.73 |
| srgn | 0.73 |
| LOC100333951 | 0.73 |
Gene Ontology
Download
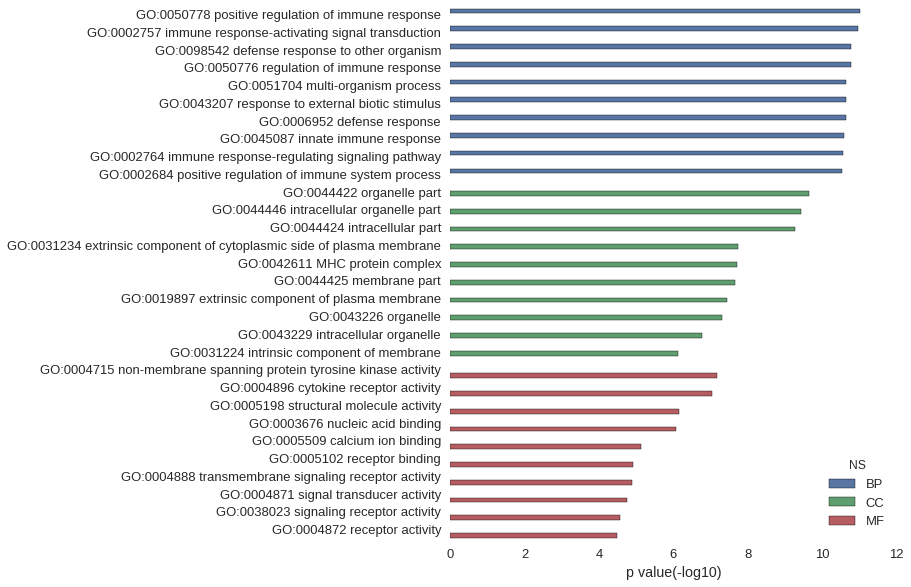
| GO | P value |
|---|---|
| GO:0050778 | 9.68e-12 |
| GO:0002757 | 1.09e-11 |
| GO:0098542 | 1.65e-11 |
| GO:0050776 | 1.68e-11 |
| GO:0051704 | 2.23e-11 |
| GO:0043207 | 2.32e-11 |
| GO:0006952 | 2.34e-11 |
| GO:0045087 | 2.57e-11 |
| GO:0002764 | 2.68e-11 |
| GO:0002684 | 2.95e-11 |
| GO:0044422 | 2.29e-10 |
| GO:0044446 | 3.80e-10 |
| GO:0044424 | 5.24e-10 |
| GO:0031234 | 1.78e-08 |
| GO:0042611 | 1.96e-08 |
| GO:0044425 | 2.20e-08 |
| GO:0019897 | 3.58e-08 |
| GO:0043226 | 4.88e-08 |
| GO:0043229 | 1.71e-07 |
| GO:0031224 | 7.65e-07 |
| GO:0004715 | 6.85e-08 |
| GO:0004896 | 9.32e-08 |
| GO:0005198 | 7.26e-07 |
| GO:0003676 | 8.26e-07 |
| GO:0005509 | 7.58e-06 |
| GO:0005102 | 1.22e-05 |
| GO:0004888 | 1.33e-05 |
| GO:0004871 | 1.82e-05 |
| GO:0038023 | 2.79e-05 |
| GO:0004872 | 3.18e-05 |
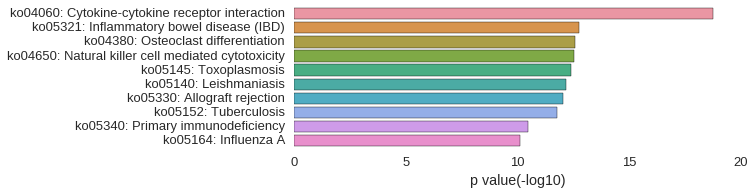
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG08586
TAGTGGCCCTAGTTATATAGTGTGTATTTATAAGTCAGTTACAAGCTTACAGCATGTATGTTTAACATGG
ACATTAAAAACAAAGAATGTCCAATCTGTGTGTTATCTGTATCATCCACTTAATAATGTATAAACAAAGG
AGGTAATACGGCCACAAAAGCCTGTGAATGAATTACTGCATTTCTGCTGCCACCATGTGCATCTCCGATC
TTTAAGCCAAGAGAAACGGTCAAAGGGAACCTGACAAGGTCCAACAGCAGCTCACTCTAAATATACAAAG
TGT
TAGTGGCCCTAGTTATATAGTGTGTATTTATAAGTCAGTTACAAGCTTACAGCATGTATGTTTAACATGG
ACATTAAAAACAAAGAATGTCCAATCTGTGTGTTATCTGTATCATCCACTTAATAATGTATAAACAAAGG
AGGTAATACGGCCACAAAAGCCTGTGAATGAATTACTGCATTTCTGCTGCCACCATGTGCATCTCCGATC
TTTAAGCCAAGAGAAACGGTCAAAGGGAACCTGACAAGGTCCAACAGCAGCTCACTCTAAATATACAAAG
TGT