ZFLNCG08767
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR891511 | brain | normal | 12.86 |
| SRR519732 | 7 dpf | FETOH treatment | 11.91 |
| SRR519752 | 7 dpf | VD3 treatment | 11.72 |
| SRR519727 | 6 dpf | FETOH treatment | 11.06 |
| SRR519747 | 6 dpf | VD3 treatment | 10.90 |
| SRR726541 | 5 dpf | infection with Mycobacterium marinum | 10.14 |
| SRR1048059 | pineal gland | light | 8.95 |
| SRR519737 | 2 dpf | VD3 treatment | 7.74 |
| SRR519717 | 2 dpf | FETOH treatment | 7.32 |
| SRR726540 | 5 dpf | normal | 7.03 |
Express in tissues
Correlated coding gene
Download
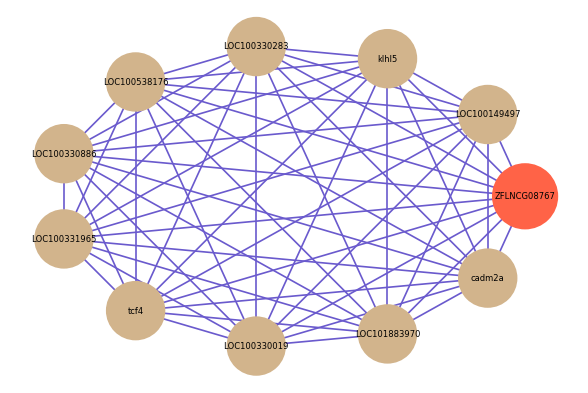
| gene | correlation coefficent |
|---|---|
| LOC100149497 | 0.74 |
| klhl5 | 0.74 |
| LOC100330283 | 0.72 |
| LOC100538176 | 0.72 |
| LOC100330886 | 0.71 |
| LOC100331965 | 0.70 |
| tcf4 | 0.70 |
| LOC100330019 | 0.69 |
| LOC101883970 | 0.69 |
| cadm2a | 0.69 |
Gene Ontology
Download
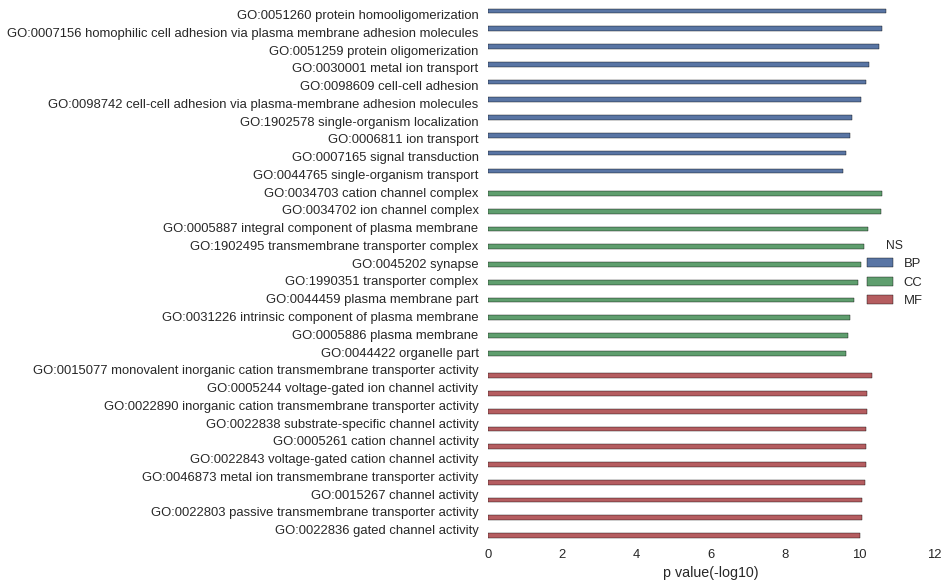
| GO | P value |
|---|---|
| GO:0051260 | 1.96e-11 |
| GO:0007156 | 2.51e-11 |
| GO:0051259 | 3.01e-11 |
| GO:0030001 | 5.49e-11 |
| GO:0098609 | 6.71e-11 |
| GO:0098742 | 9.20e-11 |
| GO:1902578 | 1.65e-10 |
| GO:0006811 | 1.78e-10 |
| GO:0007165 | 2.38e-10 |
| GO:0044765 | 2.76e-10 |
| GO:0034703 | 2.55e-11 |
| GO:0034702 | 2.69e-11 |
| GO:0005887 | 5.99e-11 |
| GO:1902495 | 7.59e-11 |
| GO:0045202 | 9.20e-11 |
| GO:1990351 | 1.12e-10 |
| GO:0044459 | 1.41e-10 |
| GO:0031226 | 1.87e-10 |
| GO:0005886 | 2.10e-10 |
| GO:0044422 | 2.36e-10 |
| GO:0015077 | 4.57e-11 |
| GO:0005244 | 6.23e-11 |
| GO:0022890 | 6.36e-11 |
| GO:0022838 | 6.65e-11 |
| GO:0005261 | 6.71e-11 |
| GO:0022843 | 6.97e-11 |
| GO:0046873 | 7.35e-11 |
| GO:0015267 | 8.57e-11 |
| GO:0022803 | 8.57e-11 |
| GO:0022836 | 9.94e-11 |
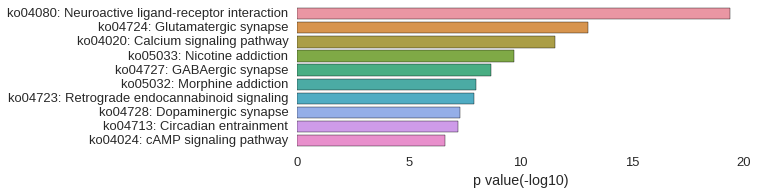
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG08767
ACACCATATAGATCAAGTTGATAGACAGAACACTGTATAGAGCCAGATGGCAAACAGATAGAACCCTATA
TAGAGCCAGTATACTGACAGTACTCTACATAGAATAAGATGAGAGACCCAATAATGTCAAATGACAGATA
ACCAGACTGACTATATCAACAGACAGACAGATGGAGAGAGATAGAGAGAGAAAGAGAGACAGACAGGTGT
GTCGTACCTGTAAGGAGTGTAAGACAGCAGCAGGTGTGTCGTCCGCTCTCAGGAACACTTGCTGTCCACG
TCTGAAACACAAACCAAAAGCATCAGAATGTGTGTGTGTGTGTCTTCACTTCTGCCCAACTCTGCAGAAG
TGTCTGTCTCTCACACATACTTCTATTACCTACTGTCCTGCGTGTGTGTGAGTGTGTGAGTGTGTGTGTG
TGTGTGTGTGAGTGTGTGAGTGTGTGTGTGTGAGTGAGTGTGTGAGTGTGTGTGTGTGTGTGGAGTGTAA
ACACGTCCACATGCAGGCGGTGTGTCTTACAGGTCTCTTCCTCGCTTCCTCTCATGAGCGTTCATGTCAT
TCCCTCGAGTGTGTCAAAGCTCCACACACACACTCTCACACACACACTCACACACACACACACTGGTGCC
GGTCTCAGAGAGGTGTGTGTGCGCTTGGGTCCTCAGCAGGAACACACTGACAGCACACGCTCAGCACACA
CACTCCTCTGTCCTCTCTCTCTCTCTTTCTCTCCCTATTGACTGGGAAGACT
ACACCATATAGATCAAGTTGATAGACAGAACACTGTATAGAGCCAGATGGCAAACAGATAGAACCCTATA
TAGAGCCAGTATACTGACAGTACTCTACATAGAATAAGATGAGAGACCCAATAATGTCAAATGACAGATA
ACCAGACTGACTATATCAACAGACAGACAGATGGAGAGAGATAGAGAGAGAAAGAGAGACAGACAGGTGT
GTCGTACCTGTAAGGAGTGTAAGACAGCAGCAGGTGTGTCGTCCGCTCTCAGGAACACTTGCTGTCCACG
TCTGAAACACAAACCAAAAGCATCAGAATGTGTGTGTGTGTGTCTTCACTTCTGCCCAACTCTGCAGAAG
TGTCTGTCTCTCACACATACTTCTATTACCTACTGTCCTGCGTGTGTGTGAGTGTGTGAGTGTGTGTGTG
TGTGTGTGTGAGTGTGTGAGTGTGTGTGTGTGAGTGAGTGTGTGAGTGTGTGTGTGTGTGTGGAGTGTAA
ACACGTCCACATGCAGGCGGTGTGTCTTACAGGTCTCTTCCTCGCTTCCTCTCATGAGCGTTCATGTCAT
TCCCTCGAGTGTGTCAAAGCTCCACACACACACTCTCACACACACACTCACACACACACACACTGGTGCC
GGTCTCAGAGAGGTGTGTGTGCGCTTGGGTCCTCAGCAGGAACACACTGACAGCACACGCTCAGCACACA
CACTCCTCTGTCCTCTCTCTCTCTCTTTCTCTCCCTATTGACTGGGAAGACT