ZFLNCG08892
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR514030 | retina | id2a morpholino | 40.68 |
| SRR514028 | retina | control morpholino | 21.30 |
| SRR594771 | endothelium | normal | 12.53 |
| SRR519717 | 2 dpf | FETOH treatment | 10.66 |
| SRR519737 | 2 dpf | VD3 treatment | 10.50 |
| SRR776509 | embryo | Salmonella infected | 8.99 |
| SRR776507 | embryo | normal | 8.22 |
| SRR1104058 | heart | normal | 8.04 |
| SRR1104059 | heart | kctd10 mut | 7.85 |
| SRR065196 | 3 dpf | normal | 7.63 |
Express in tissues
Correlated coding gene
Download
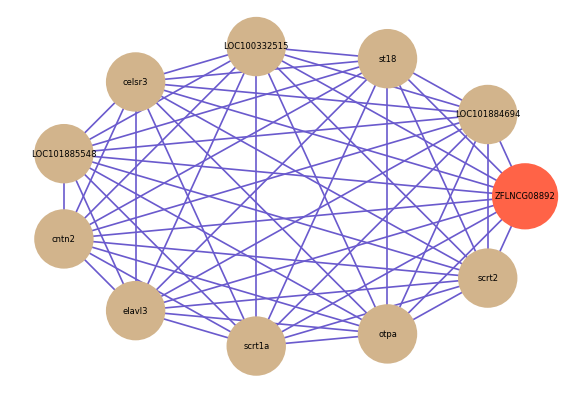
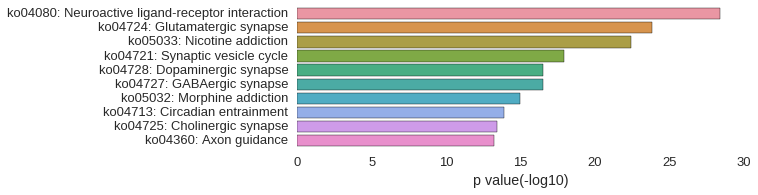
| gene | correlation coefficent |
|---|---|
| LOC101884694 | 0.90 |
| st18 | 0.89 |
| LOC100332515 | 0.88 |
| celsr3 | 0.87 |
| LOC101885548 | 0.87 |
| cntn2 | 0.87 |
| elavl3 | 0.87 |
| scrt1a | 0.87 |
| otpa | 0.87 |
| scrt2 | 0.86 |
Gene Ontology
Download
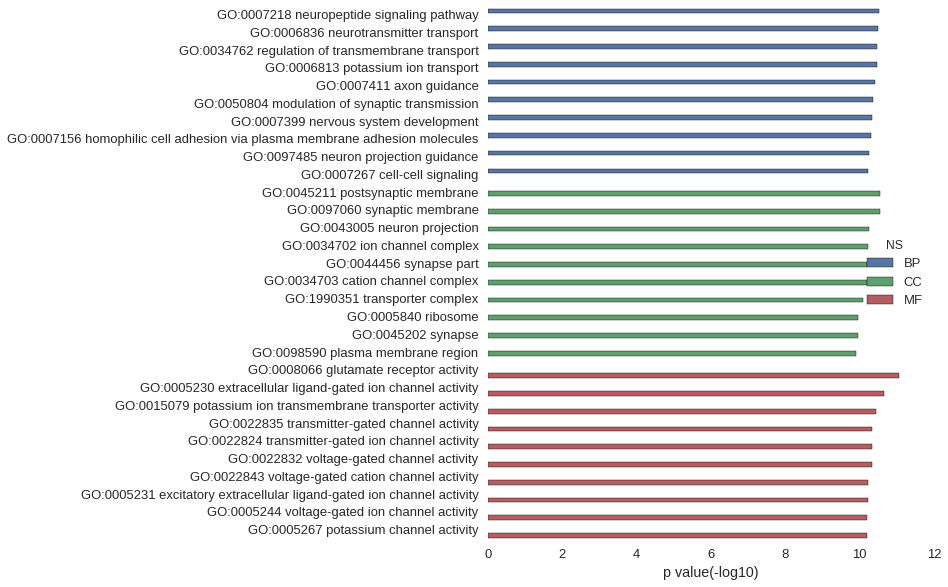
| GO | P value |
|---|---|
| GO:0007218 | 3.07e-11 |
| GO:0006836 | 3.23e-11 |
| GO:0034762 | 3.35e-11 |
| GO:0006813 | 3.35e-11 |
| GO:0007411 | 3.86e-11 |
| GO:0050804 | 4.33e-11 |
| GO:0007399 | 4.60e-11 |
| GO:0007156 | 5.06e-11 |
| GO:0097485 | 5.48e-11 |
| GO:0007267 | 5.82e-11 |
| GO:0045211 | 2.80e-11 |
| GO:0097060 | 2.86e-11 |
| GO:0043005 | 5.53e-11 |
| GO:0034702 | 5.88e-11 |
| GO:0044456 | 6.23e-11 |
| GO:0034703 | 6.47e-11 |
| GO:1990351 | 8.35e-11 |
| GO:0005840 | 1.08e-10 |
| GO:0045202 | 1.09e-10 |
| GO:0098590 | 1.26e-10 |
| GO:0008066 | 9.02e-12 |
| GO:0005230 | 2.27e-11 |
| GO:0015079 | 3.63e-11 |
| GO:0022835 | 4.60e-11 |
| GO:0022824 | 4.60e-11 |
| GO:0022832 | 4.70e-11 |
| GO:0022843 | 5.95e-11 |
| GO:0005231 | 6.13e-11 |
| GO:0005244 | 6.23e-11 |
| GO:0005267 | 6.53e-11 |
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG08892
AAGAAGAGAAGCAGGCCAGCTGAACATATACAGTGTTCAGTATCGACAAGGACAGAAATAACACTATAGT
TGAGAACCGAGATATGCAGTTATATACGCTCGCAAACATGGTAAATACATGCCTTTGAGTGACAGATACC
TATATGTTCGACAAGCAGATATACCAAATACTGTAGCTTAATACCAAAACGAACAGACAAACGTGAAAGT
ATCAGTGGAATAACCAGCGTTTGTCTTGTTTCAATCCGTCTATCCGCCTGTGTGTGCGCGCGCGCTGGGA
AAACACAGTCGCGTCTGCGTATCTATGCATGTTCATCCAAAGTACTACAAAAGAATAAGTGGTTTTTATA
AAATGTAACTGCGCTATTTAGTGAATAACCCGTTTGTGTCAGAAACCCCTGAACCACCGACGAAACCAAA
TGATTTAGCCAAGTGCGAAAAAGCCACAACTGCTCAAATTGCAAAGAAAAACTCAAATCAACAGCAAACA
GAACATCTGAAAAGAACCAGACAAAAAGACGGATCATGTTTGTTCCTTTGCCGGTGCCGTTTGTCTTGCC
GAGAACTGCCTCCGACCTCCACGCCTGGCGTCTCAAGTCCCCGCTGGCTCTCCGCTCACACTCCGGTAAG
GCTTTCTTCCACAACAATCACCTATTATTTCATTTATTTCCAAATCCGGCAAAGCATCCCATCTCCATCG
CCCTCATCATCCTTATCCCTGTTTCCACCGCGGGAATGAAGCATCCACATTTGGCGTTTTCTGTCATGTG
TCGCGCGCTGGCATTGCCGCCCTTTCCAACATTTAGGTTTAGTCCAACCTTCAAGTTTGTTTACAGCAGT
ACCTGCACTGTTAAAAGCGGCTCAGTTCGGGGTTGTTTTCTGTATAAATTTCCGTCAGGTTAGGCTCAGT
TCTTACAGGGCTCCGTCTCCGTCCGTCTCCTCCTCTCGCCTCTGCCTCTGCCTCCGTCTCCCCCGAACTA
AACTCGCGGTGAGCTGGGAATATTCATGCCAACTTATCGATCTCCGCAGGATCGATTCCGGGGCCGGCGG
ATCGAG
AAGAAGAGAAGCAGGCCAGCTGAACATATACAGTGTTCAGTATCGACAAGGACAGAAATAACACTATAGT
TGAGAACCGAGATATGCAGTTATATACGCTCGCAAACATGGTAAATACATGCCTTTGAGTGACAGATACC
TATATGTTCGACAAGCAGATATACCAAATACTGTAGCTTAATACCAAAACGAACAGACAAACGTGAAAGT
ATCAGTGGAATAACCAGCGTTTGTCTTGTTTCAATCCGTCTATCCGCCTGTGTGTGCGCGCGCGCTGGGA
AAACACAGTCGCGTCTGCGTATCTATGCATGTTCATCCAAAGTACTACAAAAGAATAAGTGGTTTTTATA
AAATGTAACTGCGCTATTTAGTGAATAACCCGTTTGTGTCAGAAACCCCTGAACCACCGACGAAACCAAA
TGATTTAGCCAAGTGCGAAAAAGCCACAACTGCTCAAATTGCAAAGAAAAACTCAAATCAACAGCAAACA
GAACATCTGAAAAGAACCAGACAAAAAGACGGATCATGTTTGTTCCTTTGCCGGTGCCGTTTGTCTTGCC
GAGAACTGCCTCCGACCTCCACGCCTGGCGTCTCAAGTCCCCGCTGGCTCTCCGCTCACACTCCGGTAAG
GCTTTCTTCCACAACAATCACCTATTATTTCATTTATTTCCAAATCCGGCAAAGCATCCCATCTCCATCG
CCCTCATCATCCTTATCCCTGTTTCCACCGCGGGAATGAAGCATCCACATTTGGCGTTTTCTGTCATGTG
TCGCGCGCTGGCATTGCCGCCCTTTCCAACATTTAGGTTTAGTCCAACCTTCAAGTTTGTTTACAGCAGT
ACCTGCACTGTTAAAAGCGGCTCAGTTCGGGGTTGTTTTCTGTATAAATTTCCGTCAGGTTAGGCTCAGT
TCTTACAGGGCTCCGTCTCCGTCCGTCTCCTCCTCTCGCCTCTGCCTCTGCCTCCGTCTCCCCCGAACTA
AACTCGCGGTGAGCTGGGAATATTCATGCCAACTTATCGATCTCCGCAGGATCGATTCCGGGGCCGGCGG
ATCGAG