ZFLNCG08959
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR023145 | heart | normal | 1252.70 |
| SRR1648855 | brain | normal | 31.52 |
| SRR516132 | skin | male and 5 month | 24.22 |
| SRR1648854 | brain | normal | 13.94 |
| ERR594417 | retina | normal | 13.46 |
| SRR1028004 | head | normal | 10.65 |
| SRR891495 | heart | normal | 10.03 |
| SRR1562528 | eye | normal | 9.92 |
| SRR1342218 | embryo | marco morpholino | 9.19 |
| ERR594416 | retina | transgenic flk1 | 8.57 |
Express in tissues
Correlated coding gene
Download
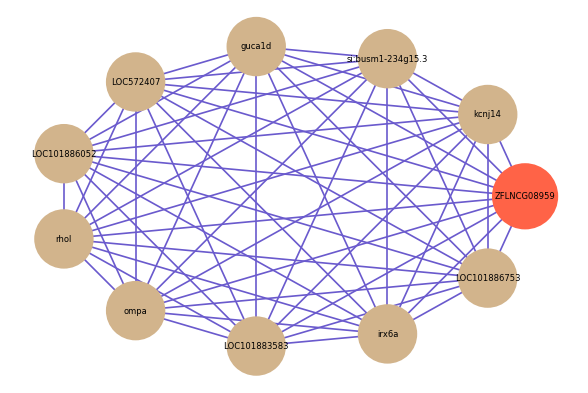
| gene | correlation coefficent |
|---|---|
| kcnj14 | 0.64 |
| si:busm1-234g15.3 | 0.59 |
| guca1d | 0.59 |
| LOC572407 | 0.59 |
| LOC101886052 | 0.59 |
| rhol | 0.59 |
| ompa | 0.58 |
| LOC101883583 | 0.58 |
| irx6a | 0.58 |
| LOC101886753 | 0.58 |
Gene Ontology
Download
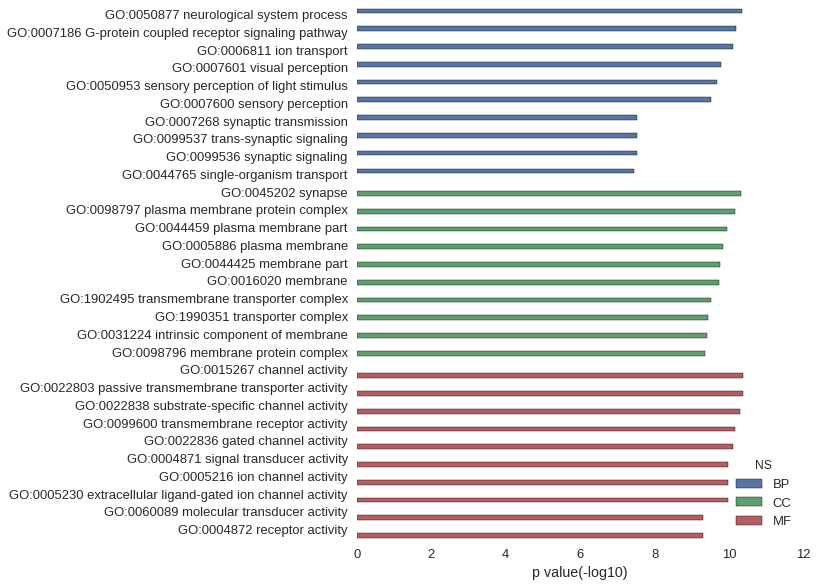
| GO | P value |
|---|---|
| GO:0050877 | 4.32e-11 |
| GO:0007186 | 6.37e-11 |
| GO:0006811 | 7.69e-11 |
| GO:0007601 | 1.68e-10 |
| GO:0050953 | 2.10e-10 |
| GO:0007600 | 2.95e-10 |
| GO:0007268 | 2.95e-08 |
| GO:0099537 | 2.95e-08 |
| GO:0099536 | 2.95e-08 |
| GO:0044765 | 3.64e-08 |
| GO:0045202 | 4.69e-11 |
| GO:0098797 | 6.67e-11 |
| GO:0044459 | 1.14e-10 |
| GO:0005886 | 1.41e-10 |
| GO:0044425 | 1.74e-10 |
| GO:0016020 | 1.89e-10 |
| GO:1902495 | 2.95e-10 |
| GO:1990351 | 3.70e-10 |
| GO:0031224 | 3.84e-10 |
| GO:0098796 | 4.51e-10 |
| GO:0015267 | 4.18e-11 |
| GO:0022803 | 4.18e-11 |
| GO:0022838 | 4.94e-11 |
| GO:0099600 | 7.02e-11 |
| GO:0022836 | 7.56e-11 |
| GO:0004871 | 1.03e-10 |
| GO:0005216 | 1.04e-10 |
| GO:0005230 | 1.08e-10 |
| GO:0060089 | 4.83e-10 |
| GO:0004872 | 4.83e-10 |
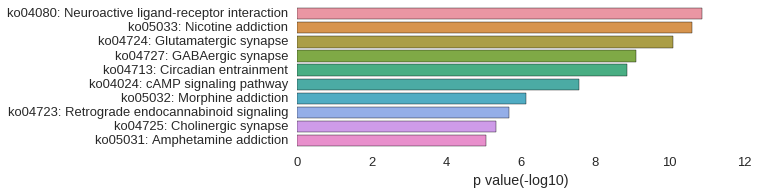
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG08959
ATTTGTAAAGTACGTTCAAAATAATTCAAAATGCATTACCTAGGTCTACTTATGTGATTTCAGATGCATA
AAAGTTGACCAAGTTGGCTAAGCGAGATGTCCTTTACTACAGCCAAGATCTTGGAGATTTTATGCTTTCT
TAATCAAAGTATTTACTGAAACAGCATGGGGCTATTAACATGCCAAAACAATCTGAATTTAAATTGTGTT
AATTTGAATTATTAACTGTTGGAATTTAATAAAAC
ATTTGTAAAGTACGTTCAAAATAATTCAAAATGCATTACCTAGGTCTACTTATGTGATTTCAGATGCATA
AAAGTTGACCAAGTTGGCTAAGCGAGATGTCCTTTACTACAGCCAAGATCTTGGAGATTTTATGCTTTCT
TAATCAAAGTATTTACTGAAACAGCATGGGGCTATTAACATGCCAAAACAATCTGAATTTAAATTGTGTT
AATTTGAATTATTAACTGTTGGAATTTAATAAAAC