ZFLNCG08960
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR023145 | heart | normal | 570.67 |
| ERR594417 | retina | normal | 11.22 |
| SRR891495 | heart | normal | 8.06 |
| ERR594416 | retina | transgenic flk1 | 7.41 |
| SRR1562528 | eye | normal | 5.77 |
| SRR1648854 | brain | normal | 4.65 |
| SRR957181 | heart | normal | 4.25 |
| SRR527834 | head | normal | 3.18 |
| SRR630464 | 5 dpf | injected epidermidis | 3.09 |
| SRR1648855 | brain | normal | 2.88 |
Express in tissues
Correlated coding gene
Download
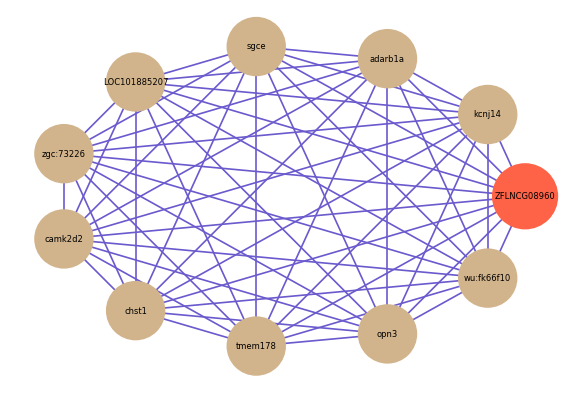
| gene | correlation coefficent |
|---|---|
| kcnj14 | 0.74 |
| adarb1a | 0.69 |
| sgce | 0.68 |
| LOC101885207 | 0.68 |
| zgc:73226 | 0.68 |
| camk2d2 | 0.67 |
| chst1 | 0.67 |
| tmem178 | 0.67 |
| opn3 | 0.67 |
| wu:fk66f10 | 0.67 |
Gene Ontology
Download
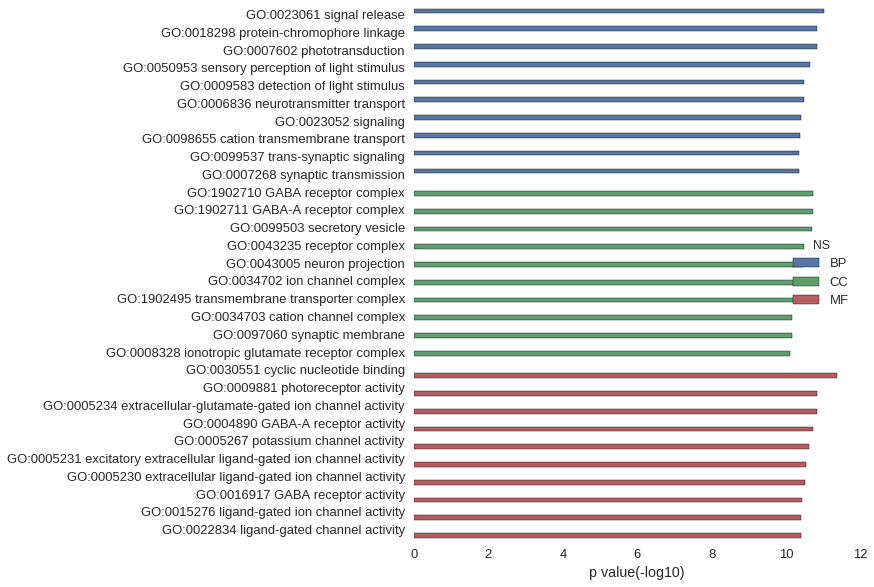
| GO | P value |
|---|---|
| GO:0023061 | 9.63e-12 |
| GO:0018298 | 1.44e-11 |
| GO:0007602 | 1.44e-11 |
| GO:0050953 | 2.33e-11 |
| GO:0009583 | 3.38e-11 |
| GO:0006836 | 3.39e-11 |
| GO:0023052 | 4.01e-11 |
| GO:0098655 | 4.26e-11 |
| GO:0099537 | 4.50e-11 |
| GO:0007268 | 4.50e-11 |
| GO:1902710 | 1.92e-11 |
| GO:1902711 | 1.92e-11 |
| GO:0099503 | 2.04e-11 |
| GO:0043235 | 3.36e-11 |
| GO:0043005 | 3.40e-11 |
| GO:0034702 | 3.94e-11 |
| GO:1902495 | 5.45e-11 |
| GO:0034703 | 6.92e-11 |
| GO:0097060 | 7.12e-11 |
| GO:0008328 | 7.75e-11 |
| GO:0030551 | 4.27e-12 |
| GO:0009881 | 1.44e-11 |
| GO:0005234 | 1.49e-11 |
| GO:0004890 | 1.92e-11 |
| GO:0005267 | 2.35e-11 |
| GO:0005231 | 2.87e-11 |
| GO:0005230 | 3.18e-11 |
| GO:0016917 | 3.75e-11 |
| GO:0015276 | 3.89e-11 |
| GO:0022834 | 3.89e-11 |
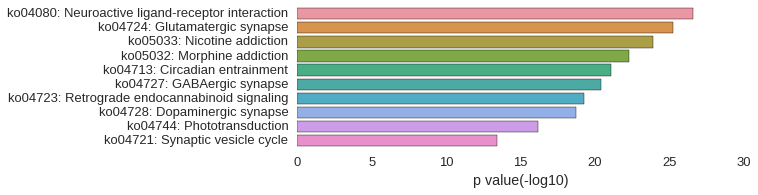
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG08960
CAAGATATGTATTAAATGATATAAAGCACAGAAGATTTTTGTGACTTTGTGAATTGTGGATTTTTCAGTT
CCATGAAAATATGTGAATATTTATGCATGCTGTTTTTAGTTATGTGGAGGCGAAATCTGGCCTAGTTACA
GCTTGTGTCACTTAAAGCAATATTTTACCATACAAATGAGAGTTCTGTCATCATGTACTTGCTGTTCTAG
ACCTAATGCTCAGCATATATAAGTACACCCCATGCAAATCCATCTTTTAAATTCATGTTTTAATCAGAAG
CTATACAATATTTAATTTGTACATATACGTTATATTTGTCAGAACTGAAGCCAAATCTGGAGCTTATCTA
ACAAAATAACTCACGATTAACGGTCAAAAAAAACTAGTACAGTGCACCC
CAAGATATGTATTAAATGATATAAAGCACAGAAGATTTTTGTGACTTTGTGAATTGTGGATTTTTCAGTT
CCATGAAAATATGTGAATATTTATGCATGCTGTTTTTAGTTATGTGGAGGCGAAATCTGGCCTAGTTACA
GCTTGTGTCACTTAAAGCAATATTTTACCATACAAATGAGAGTTCTGTCATCATGTACTTGCTGTTCTAG
ACCTAATGCTCAGCATATATAAGTACACCCCATGCAAATCCATCTTTTAAATTCATGTTTTAATCAGAAG
CTATACAATATTTAATTTGTACATATACGTTATATTTGTCAGAACTGAAGCCAAATCTGGAGCTTATCTA
ACAAAATAACTCACGATTAACGGTCAAAAAAAACTAGTACAGTGCACCC