ZFLNCG09003
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR797917 | embryo | pbx2/4-MO and prdm1-/- | 26.37 |
| SRR797908 | embryo | normal | 22.20 |
| ERR023144 | brain | normal | 19.78 |
| SRR797914 | embryo | pbx2/4-MO | 19.35 |
| SRR797911 | embryo | prdm1-/- | 19.28 |
| SRR1565805 | brain | normal | 7.93 |
| SRR1565807 | brain | normal | 7.60 |
| SRR1565809 | brain | normal | 7.11 |
| SRR1565813 | brain | normal | 6.88 |
| SRR1565815 | brain | normal | 6.80 |
Express in tissues
Correlated coding gene
Download
Gene Ontology
Download
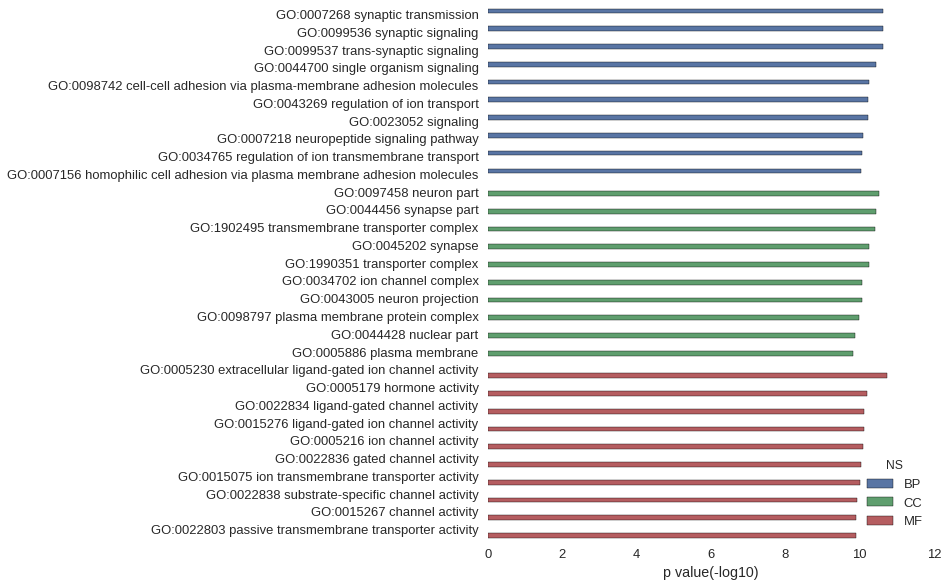
| GO | P value |
|---|---|
| GO:0007268 | 2.41e-11 |
| GO:0099536 | 2.41e-11 |
| GO:0099537 | 2.41e-11 |
| GO:0044700 | 3.65e-11 |
| GO:0098742 | 5.66e-11 |
| GO:0043269 | 5.94e-11 |
| GO:0023052 | 6.06e-11 |
| GO:0007218 | 8.20e-11 |
| GO:0034765 | 8.78e-11 |
| GO:0007156 | 9.05e-11 |
| GO:0097458 | 3.02e-11 |
| GO:0044456 | 3.53e-11 |
| GO:1902495 | 3.91e-11 |
| GO:0045202 | 5.66e-11 |
| GO:1990351 | 5.70e-11 |
| GO:0034702 | 8.47e-11 |
| GO:0043005 | 8.73e-11 |
| GO:0098797 | 1.02e-10 |
| GO:0044428 | 1.30e-10 |
| GO:0005886 | 1.48e-10 |
| GO:0005230 | 1.89e-11 |
| GO:0005179 | 6.45e-11 |
| GO:0022834 | 7.51e-11 |
| GO:0015276 | 7.51e-11 |
| GO:0005216 | 7.98e-11 |
| GO:0022836 | 9.12e-11 |
| GO:0015075 | 1.01e-10 |
| GO:0022838 | 1.20e-10 |
| GO:0015267 | 1.28e-10 |
| GO:0022803 | 1.28e-10 |
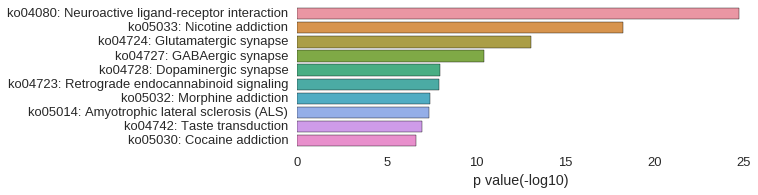
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG09003
TTGGTTTTATAAAGACTTGCATGATGGTTCCAAACACAATCAATAAACAGTGTGAAGTATAACAGTAATG
TACAGTAAAATGATCACACAATAAGTTATTTTAACAGTTTTCCAGTGCTTTCATGCAATGCTAAAGAAAC
AACAGCAAAAGATCACAGTGATATCTATTTATACATTCATCAAACAAACGCTCAGTTTCTGCTTTGCAAG
TATACGGTAGGAATCCCGTTAGCAACTTTGTTTCAAATGAACGCCAGGCAGCAGCACAAACTCACAGATA
AACATCTGAGCACTGTAAACACTTCATAAATTGATAAATTCAGTGTTTTTATCTGAATATCTGCACATAT
TCACATATTTTTTTGTCACATATGCCATTTTGTTGTTGTTTTTGGACAAAATCTCAAAAATATGGGTTCT
AAATAT
TTGGTTTTATAAAGACTTGCATGATGGTTCCAAACACAATCAATAAACAGTGTGAAGTATAACAGTAATG
TACAGTAAAATGATCACACAATAAGTTATTTTAACAGTTTTCCAGTGCTTTCATGCAATGCTAAAGAAAC
AACAGCAAAAGATCACAGTGATATCTATTTATACATTCATCAAACAAACGCTCAGTTTCTGCTTTGCAAG
TATACGGTAGGAATCCCGTTAGCAACTTTGTTTCAAATGAACGCCAGGCAGCAGCACAAACTCACAGATA
AACATCTGAGCACTGTAAACACTTCATAAATTGATAAATTCAGTGTTTTTATCTGAATATCTGCACATAT
TCACATATTTTTTTGTCACATATGCCATTTTGTTGTTGTTTTTGGACAAAATCTCAAAAATATGGGTTCT
AAATAT