ZFLNCG09399
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR145646 | skeletal muscle | 32 degree_C to 27 degree_C | 20.80 |
| SRR516132 | skin | male and 5 month | 15.20 |
| ERR145648 | skeletal muscle | 27 degree_C to 27 degree_C | 14.56 |
| SRR801555 | embryo | RPS19 morpholino | 12.69 |
| SRR516130 | skin | male and 5 month | 10.55 |
| SRR1291414 | 5 dpi | normal | 9.90 |
| ERR145635 | skeletal muscle | 27 degree_C to 16 degree_C | 9.67 |
| SRR800045 | muscle | normal | 8.58 |
| SRR801556 | embryo | RPS19 and p53 morpholino | 7.72 |
| ERR145631 | skeletal muscle | 32 degree_C to 16 degree_C | 7.66 |
Express in tissues
Correlated coding gene
Download
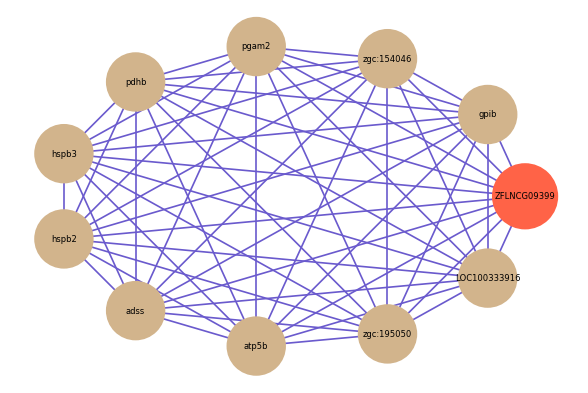
| gene | correlation coefficent |
|---|---|
| gpib | 0.63 |
| zgc:154046 | 0.61 |
| pgam2 | 0.60 |
| pdhb | 0.59 |
| hspb3 | 0.59 |
| hspb2 | 0.59 |
| adss | 0.58 |
| atp5b | 0.58 |
| zgc:195050 | 0.58 |
| LOC100333916 | 0.57 |
Gene Ontology
Download
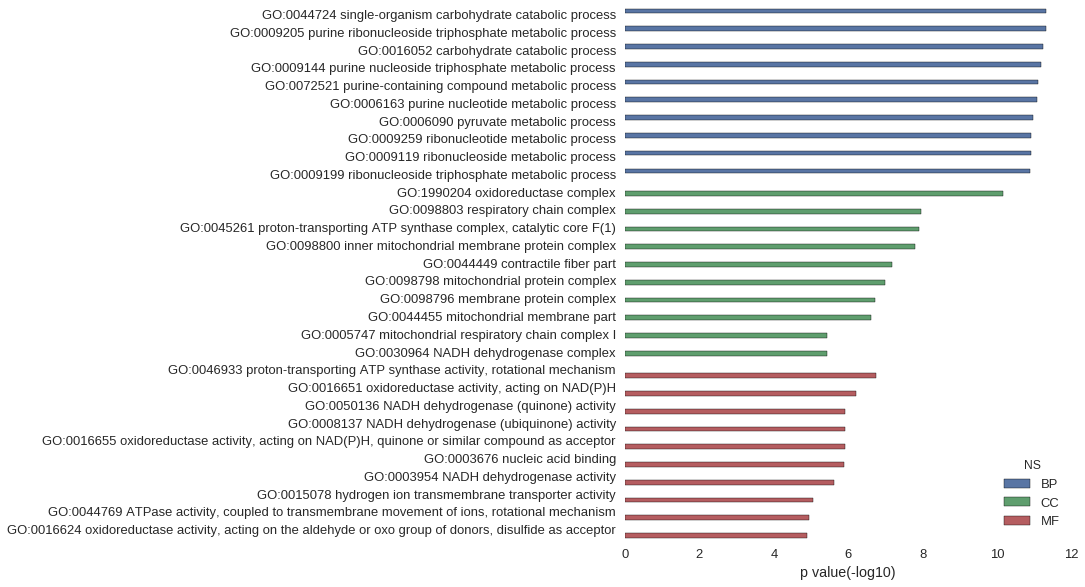
| GO | P value |
|---|---|
| GO:0044724 | 4.69e-12 |
| GO:0009205 | 4.79e-12 |
| GO:0016052 | 5.69e-12 |
| GO:0009144 | 6.48e-12 |
| GO:0072521 | 7.83e-12 |
| GO:0006163 | 8.19e-12 |
| GO:0006090 | 1.11e-11 |
| GO:0009259 | 1.21e-11 |
| GO:0009119 | 1.21e-11 |
| GO:0009199 | 1.33e-11 |
| GO:1990204 | 6.89e-11 |
| GO:0098803 | 1.14e-08 |
| GO:0045261 | 1.29e-08 |
| GO:0098800 | 1.57e-08 |
| GO:0044449 | 6.60e-08 |
| GO:0098798 | 1.00e-07 |
| GO:0098796 | 1.87e-07 |
| GO:0044455 | 2.41e-07 |
| GO:0005747 | 3.66e-06 |
| GO:0030964 | 3.66e-06 |
| GO:0046933 | 1.78e-07 |
| GO:0016651 | 6.13e-07 |
| GO:0050136 | 1.23e-06 |
| GO:0008137 | 1.23e-06 |
| GO:0016655 | 1.23e-06 |
| GO:0003676 | 1.29e-06 |
| GO:0003954 | 2.46e-06 |
| GO:0015078 | 8.73e-06 |
| GO:0044769 | 1.15e-05 |
| GO:0016624 | 1.27e-05 |
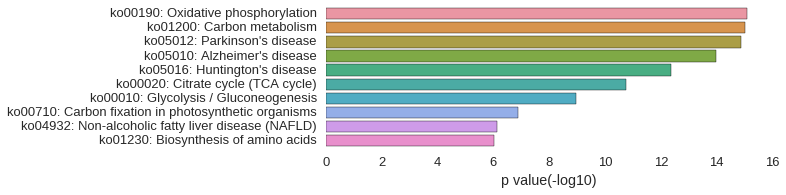
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG09399
CAGGGATGACGGATCTTTCACAGTCAGCAATGTCCCGTCTGGATCATATGTTCTGGAAATCTCCTCACCG
ACCTACAGATTCCTGCCGGTGCGAGTGGACATCACCTCCGCAGGGAAGATGAGGTGTGTGTGTGTGTGTG
AGGTATTGACTGCAGTCCTGCGGGACCAGGTCTGATGGATGAATGCGGGTGCGGGCGATGAGGTCCTCCT
CTGTGCACTCAAAATATGGCTTTTGCTGCTTGCTTAAATGATCTTTGATTCTTTTGTGAGCCCTGTGTCA
GAATAAAGCCATGTTGGTCAATTAAAGGAGAGAAGTGCGCATACTGACGCTTCTGAAAATCTTTCGTACA
TGTGGACATAGCCTGAGTCAGTCCTGATGGTGTGTGTGTGTGTGTGTGCAGGGCTCGTGTGGTGAACTAC
ATCCAGACGTCAGAGGTGATCCTGCTGCCGTACCCGCTGCAGATGCGGAGTGTTGGTCTGCACCGATTCT
TCACGGAGCGCGAGAGCTGGGGATGGACAGACTTCATCCTCAACCCCATGGTGCGCTACTTTAATTCATT
ACATACACAGTTATCAAAGTCAGAATTATCAGCCCTCTTTTATTTCTTCTAATCATAATAGTTTTAATAA
CTCATCTCTAATAACTGATGTCTTTTCTCTTGGTCAGGATGACAGCACATAATATTAGACTAGATATTCT
TCAAGACACTTATATACAGCTTAAAGCCACATTAGTAATTCATTATTTTTTGTAGTGACTTACCCAACAC
TTTTGTCATGTTCTGGTAGGTGCTCATGATGGTTCTTCCTCTCCTCGTCATCCTCCTCCTCCCGAGAGTC
TTCAACCCCAGCGATCCAGAGATGAGACGGG
CAGGGATGACGGATCTTTCACAGTCAGCAATGTCCCGTCTGGATCATATGTTCTGGAAATCTCCTCACCG
ACCTACAGATTCCTGCCGGTGCGAGTGGACATCACCTCCGCAGGGAAGATGAGGTGTGTGTGTGTGTGTG
AGGTATTGACTGCAGTCCTGCGGGACCAGGTCTGATGGATGAATGCGGGTGCGGGCGATGAGGTCCTCCT
CTGTGCACTCAAAATATGGCTTTTGCTGCTTGCTTAAATGATCTTTGATTCTTTTGTGAGCCCTGTGTCA
GAATAAAGCCATGTTGGTCAATTAAAGGAGAGAAGTGCGCATACTGACGCTTCTGAAAATCTTTCGTACA
TGTGGACATAGCCTGAGTCAGTCCTGATGGTGTGTGTGTGTGTGTGTGCAGGGCTCGTGTGGTGAACTAC
ATCCAGACGTCAGAGGTGATCCTGCTGCCGTACCCGCTGCAGATGCGGAGTGTTGGTCTGCACCGATTCT
TCACGGAGCGCGAGAGCTGGGGATGGACAGACTTCATCCTCAACCCCATGGTGCGCTACTTTAATTCATT
ACATACACAGTTATCAAAGTCAGAATTATCAGCCCTCTTTTATTTCTTCTAATCATAATAGTTTTAATAA
CTCATCTCTAATAACTGATGTCTTTTCTCTTGGTCAGGATGACAGCACATAATATTAGACTAGATATTCT
TCAAGACACTTATATACAGCTTAAAGCCACATTAGTAATTCATTATTTTTTGTAGTGACTTACCCAACAC
TTTTGTCATGTTCTGGTAGGTGCTCATGATGGTTCTTCCTCTCCTCGTCATCCTCCTCCTCCCGAGAGTC
TTCAACCCCAGCGATCCAGAGATGAGACGGG