ZFLNCG09420
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR592702 | pineal gland | normal | 296.67 |
| SRR592701 | pineal gland | normal | 288.30 |
| SRR592699 | pineal gland | normal | 223.93 |
| SRR592703 | pineal gland | normal | 219.81 |
| SRR592700 | pineal gland | normal | 216.95 |
| SRR592698 | pineal gland | normal | 74.37 |
| ERR023144 | brain | normal | 65.62 |
| SRR1648854 | brain | normal | 38.11 |
| SRR1048061 | pineal gland | dark | 33.86 |
| SRR1648855 | brain | normal | 30.86 |
Express in tissues
Correlated coding gene
Download
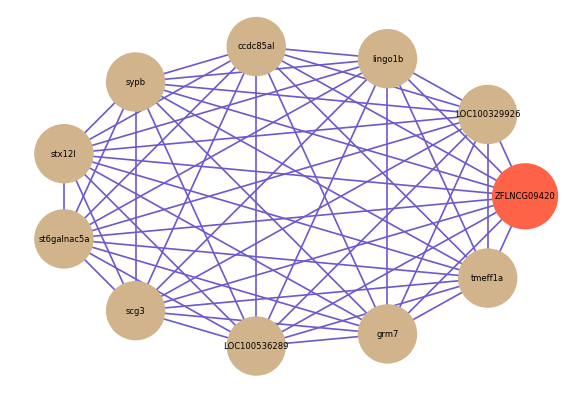
| gene | correlation coefficent |
|---|---|
| LOC100329926 | 0.87 |
| lingo1b | 0.86 |
| ccdc85al | 0.85 |
| sypb | 0.84 |
| stx12l | 0.84 |
| st6galnac5a | 0.84 |
| scg3 | 0.84 |
| LOC100536289 | 0.84 |
| grm7 | 0.84 |
| tmeff1a | 0.84 |
Gene Ontology
Download
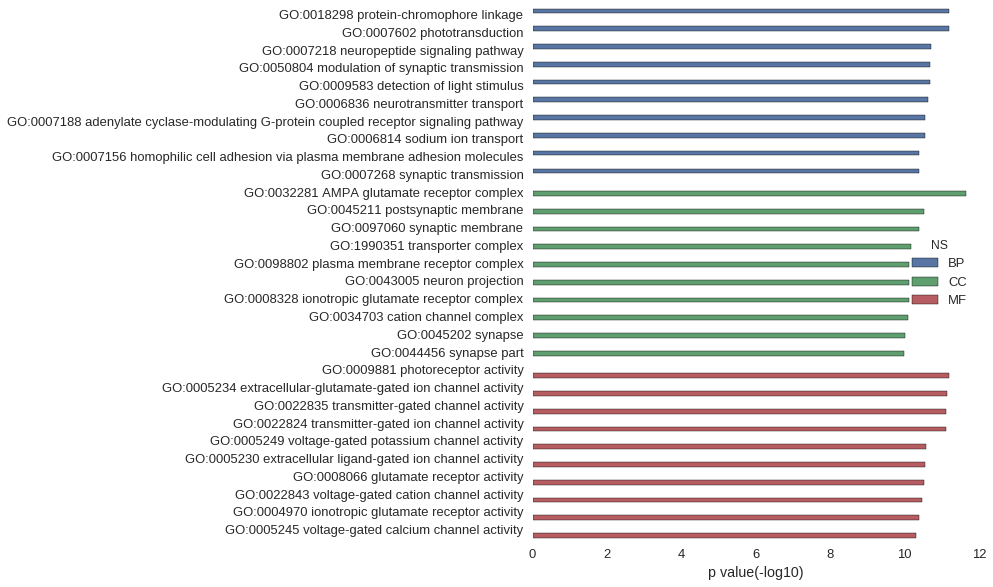
| GO | P value |
|---|---|
| GO:0018298 | 6.34e-12 |
| GO:0007602 | 6.34e-12 |
| GO:0007218 | 2.00e-11 |
| GO:0050804 | 2.03e-11 |
| GO:0009583 | 2.13e-11 |
| GO:0006836 | 2.35e-11 |
| GO:0007188 | 2.88e-11 |
| GO:0006814 | 2.88e-11 |
| GO:0007156 | 4.10e-11 |
| GO:0007268 | 4.17e-11 |
| GO:0032281 | 2.21e-12 |
| GO:0045211 | 2.93e-11 |
| GO:0097060 | 4.09e-11 |
| GO:1990351 | 6.69e-11 |
| GO:0098802 | 7.50e-11 |
| GO:0043005 | 7.50e-11 |
| GO:0008328 | 7.51e-11 |
| GO:0034703 | 8.18e-11 |
| GO:0045202 | 9.79e-11 |
| GO:0044456 | 1.02e-10 |
| GO:0009881 | 6.34e-12 |
| GO:0005234 | 7.44e-12 |
| GO:0022835 | 7.56e-12 |
| GO:0022824 | 7.56e-12 |
| GO:0005249 | 2.62e-11 |
| GO:0005230 | 2.77e-11 |
| GO:0008066 | 3.05e-11 |
| GO:0022843 | 3.31e-11 |
| GO:0004970 | 4.00e-11 |
| GO:0005245 | 5.07e-11 |
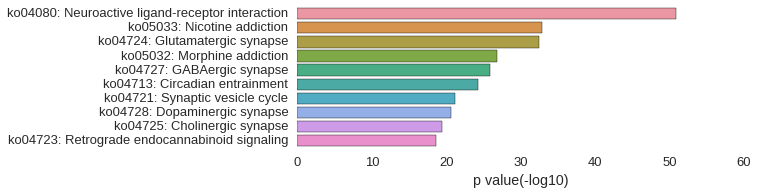
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG09420
ATTTATTCATTTCCCTTCAGCTTAGTCCCTTATTTTTCCAGGGGTTGCCACAGCGGAATGAACTGCCAAC
TATTCCGGTGTTGTGACATTTTATCCTACCCATCAGATTATGTTGAGTATACCAACACTGTATTTGAATA
GTTCTGAGGTTGGGGTTAGGGATTAGGTAGGTTAGGGCAAGATTACAGCTTCATATCACTCTACTCCACA
TTCAAATTTACGTTGGTAGCAAAATTTGATGGGTAACAAATTGAGTAATCAAAACGTGCTACCTACTTTT
AGAACAGGAGGTAGGACAATTTGACAAGC
ATTTATTCATTTCCCTTCAGCTTAGTCCCTTATTTTTCCAGGGGTTGCCACAGCGGAATGAACTGCCAAC
TATTCCGGTGTTGTGACATTTTATCCTACCCATCAGATTATGTTGAGTATACCAACACTGTATTTGAATA
GTTCTGAGGTTGGGGTTAGGGATTAGGTAGGTTAGGGCAAGATTACAGCTTCATATCACTCTACTCCACA
TTCAAATTTACGTTGGTAGCAAAATTTGATGGGTAACAAATTGAGTAATCAAAACGTGCTACCTACTTTT
AGAACAGGAGGTAGGACAATTTGACAAGC