ZFLNCG09578
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR023143 | swim bladder | normal | 75.01 |
| ERR023144 | brain | normal | 64.01 |
| ERR023145 | heart | normal | 40.55 |
| SRR372802 | 5 dpf | normal | 34.93 |
| ERR023146 | head kidney | normal | 31.03 |
| SRR1648856 | brain | normal | 28.89 |
| SRR592702 | pineal gland | normal | 28.72 |
| SRR1648854 | brain | normal | 27.73 |
| SRR516133 | skin | male and 5 month | 24.54 |
| SRR1534351 | larvae | 500 ug/l BDE47 treatment | 24.08 |
Express in tissues
Correlated coding gene
Download
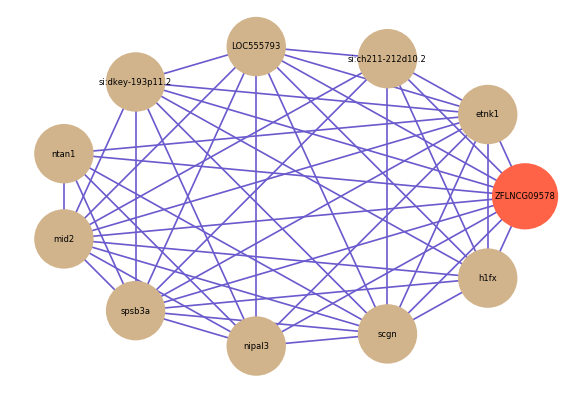
| gene | correlation coefficent |
|---|---|
| etnk1 | 0.68 |
| si:ch211-212d10.2 | 0.67 |
| LOC555793 | 0.66 |
| si:dkey-193p11.2 | 0.65 |
| ntan1 | 0.65 |
| mid2 | 0.65 |
| spsb3a | 0.65 |
| nipal3 | 0.64 |
| scgn | 0.64 |
| h1fx | 0.64 |
Gene Ontology
Download
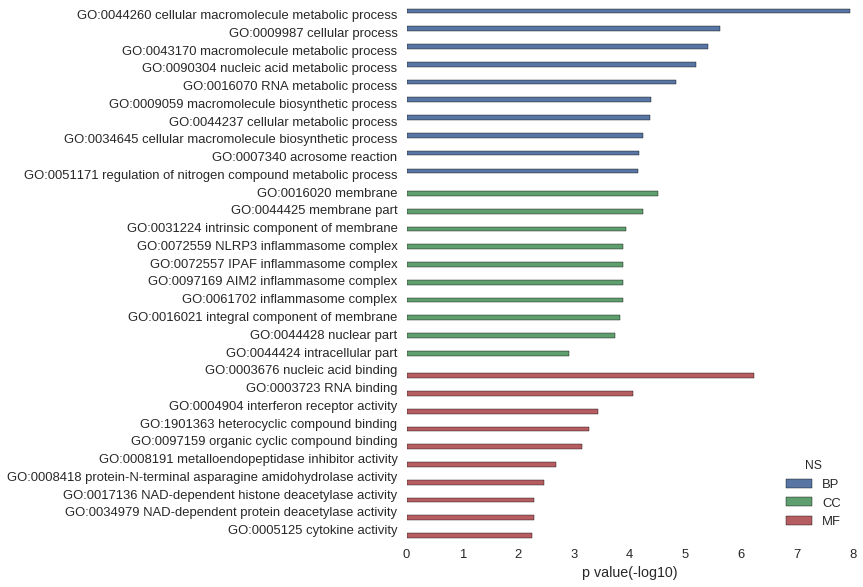
| GO | P value |
|---|---|
| GO:0044260 | 1.13e-08 |
| GO:0009987 | 2.39e-06 |
| GO:0043170 | 4.02e-06 |
| GO:0090304 | 6.45e-06 |
| GO:0016070 | 1.46e-05 |
| GO:0009059 | 4.23e-05 |
| GO:0044237 | 4.28e-05 |
| GO:0034645 | 5.86e-05 |
| GO:0007340 | 6.71e-05 |
| GO:0051171 | 6.97e-05 |
| GO:0016020 | 3.11e-05 |
| GO:0044425 | 5.70e-05 |
| GO:0031224 | 1.17e-04 |
| GO:0072559 | 1.32e-04 |
| GO:0072557 | 1.32e-04 |
| GO:0097169 | 1.32e-04 |
| GO:0061702 | 1.32e-04 |
| GO:0016021 | 1.50e-04 |
| GO:0044428 | 1.87e-04 |
| GO:0044424 | 1.24e-03 |
| GO:0003676 | 5.94e-07 |
| GO:0003723 | 8.79e-05 |
| GO:0004904 | 3.64e-04 |
| GO:1901363 | 5.35e-04 |
| GO:0097159 | 7.06e-04 |
| GO:0008191 | 2.13e-03 |
| GO:0008418 | 3.50e-03 |
| GO:0017136 | 5.19e-03 |
| GO:0034979 | 5.19e-03 |
| GO:0005125 | 5.70e-03 |
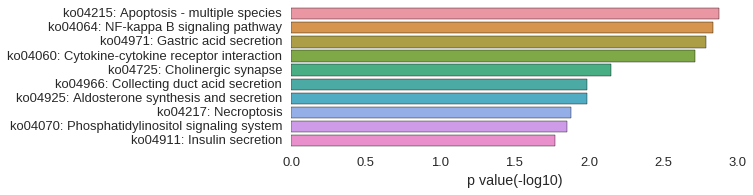
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG09578
TAATTTGTAACTTAAAAAGTCTTTTAGTTTGGAGTCTGTGCTTTTGGCTGTGGTGTAGAAACTTAAAAAG
AGATAGTTCACCCAAAAATAAAAAACTACAATTAAATTACAATAATCAATAAACAATGACTTTTTTTAGC
TCAACCTGTGGTCCTTGAGGATTCATAAAATGCAATTCCTCCATCCCCATTGTAAACTCTATCAATTCTA
TTTTGCCTTCTGAGTGCTGGTAGTTGTTAACTTACATTTTATGAATCACCAAAAACACAGTCTCAGTTGA
AAACTTCACTGCTGCACAAAAGAAAAAAATCACCAACATCTAGATTGCACAAAAG
TAATTTGTAACTTAAAAAGTCTTTTAGTTTGGAGTCTGTGCTTTTGGCTGTGGTGTAGAAACTTAAAAAG
AGATAGTTCACCCAAAAATAAAAAACTACAATTAAATTACAATAATCAATAAACAATGACTTTTTTTAGC
TCAACCTGTGGTCCTTGAGGATTCATAAAATGCAATTCCTCCATCCCCATTGTAAACTCTATCAATTCTA
TTTTGCCTTCTGAGTGCTGGTAGTTGTTAACTTACATTTTATGAATCACCAAAAACACAGTCTCAGTTGA
AAACTTCACTGCTGCACAAAAGAAAAAAATCACCAACATCTAGATTGCACAAAAG