ZFLNCG09602
Basic Information
Chromesome: chr17
Start: 25429315
End: 25430120
Transcript: ZFLNCT14799
Known as: ENSDARG00000094975
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR700534 | heart | control morpholino | 69.56 |
| SRR700537 | heart | Gata4 morpholino | 48.69 |
| SRR1205151 | 5dpf | transgenic sqET20 and neomycin treated 1h | 36.49 |
| SRR1205154 | 5dpf | transgenic sqET20 | 34.87 |
| SRR957180 | heart | 7 days after heart tip amputation | 32.99 |
| SRR594769 | blood | normal | 28.08 |
| SRR1562529 | intestine and pancreas | normal | 27.56 |
| SRR1562532 | spleen | normal | 25.61 |
| SRR1647679 | head kidney | normal | 25.15 |
| SRR957181 | heart | normal | 24.79 |
Express in tissues
Correlated coding gene
Download
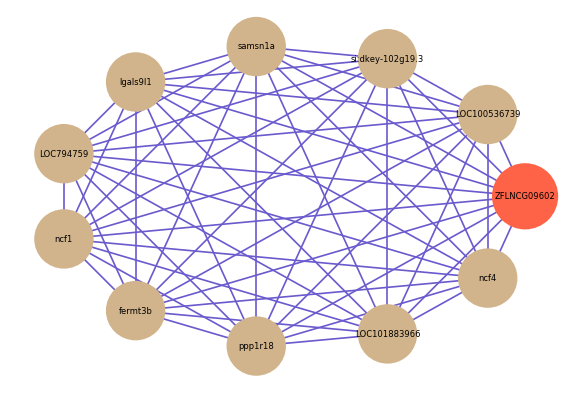
| gene | correlation coefficent |
|---|---|
| LOC100536739 | 0.72 |
| si:dkey-102g19.3 | 0.71 |
| samsn1a | 0.71 |
| lgals9l1 | 0.71 |
| LOC794759 | 0.71 |
| ncf1 | 0.71 |
| fermt3b | 0.70 |
| ppp1r18 | 0.70 |
| LOC101883966 | 0.70 |
| ncf4 | 0.70 |
Gene Ontology
Download
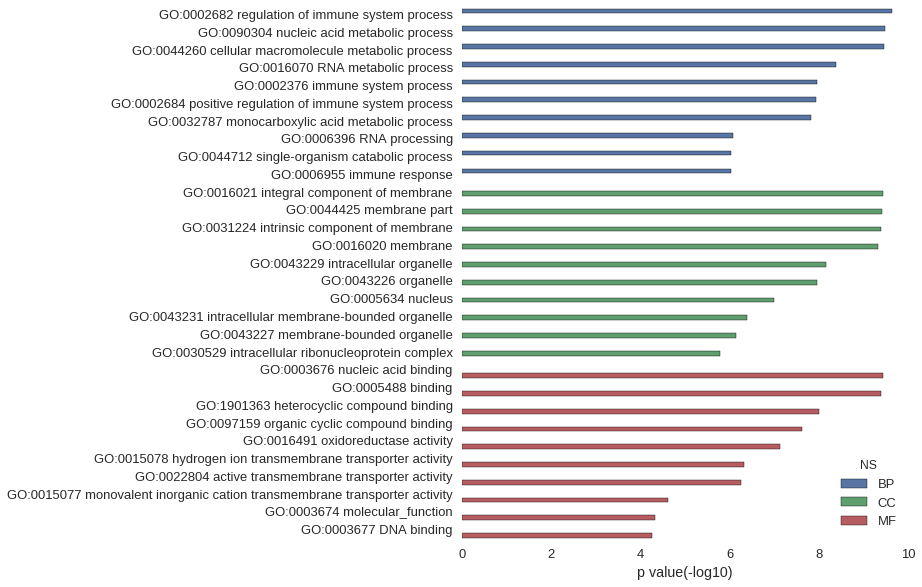
| GO | P value |
|---|---|
| GO:0002682 | 2.29e-10 |
| GO:0090304 | 3.35e-10 |
| GO:0044260 | 3.57e-10 |
| GO:0016070 | 4.15e-09 |
| GO:0002376 | 1.10e-08 |
| GO:0002684 | 1.15e-08 |
| GO:0032787 | 1.48e-08 |
| GO:0006396 | 8.48e-07 |
| GO:0044712 | 9.16e-07 |
| GO:0006955 | 9.38e-07 |
| GO:0016021 | 3.76e-10 |
| GO:0044425 | 3.81e-10 |
| GO:0031224 | 4.15e-10 |
| GO:0016020 | 4.78e-10 |
| GO:0043229 | 7.07e-09 |
| GO:0043226 | 1.10e-08 |
| GO:0005634 | 1.00e-07 |
| GO:0043231 | 4.05e-07 |
| GO:0043227 | 7.18e-07 |
| GO:0030529 | 1.65e-06 |
| GO:0003676 | 3.78e-10 |
| GO:0005488 | 4.10e-10 |
| GO:1901363 | 1.01e-08 |
| GO:0097159 | 2.42e-08 |
| GO:0016491 | 7.55e-08 |
| GO:0015078 | 4.91e-07 |
| GO:0022804 | 5.61e-07 |
| GO:0015077 | 2.42e-05 |
| GO:0003674 | 4.61e-05 |
| GO:0003677 | 5.54e-05 |
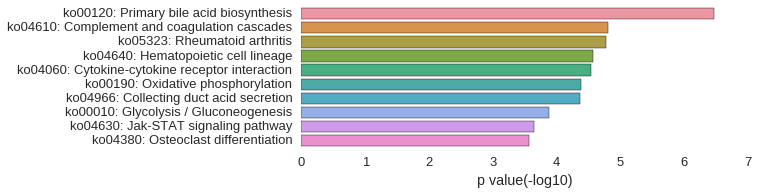
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG09602
GCATTTAGCAGTTTAATCATGCATACAGACACACTTGCCAGTCAATTACATTAATACACATAATGCCATT
AGAGAAATAGAATCTTTCAAACGCATGATTTAAGTAGTGGAGTTGTTTACCCTGCTAAACTTTATTCACA
TGAAGCAAATAGAAGAAAGGGGCTCTTCCAAGAGAAGTTGCCACTGAGCGTAACCAGACAATCCTTCTTG
GAGGAGTTCTTATTTTCATTAGTTAAAAATCCTTTGAGCTGTGCTGTGCTGTGTTCTGTCTTGCAGATTT
GTGCTCTGTCTGTCAATGACTATCTATTCTGCCACTCCCACATTGATGCTCGCCTTTGGTGAATGTTCAG
GATCTGCTGTGGATGAAGCCCAGGGTGGCAGCCGGGAACAGAAGGTACAGTGTTTTGGGCACAGGACTCA
GTCCTACAGCACTGGAGTTAGTAGATGCGCTGGGCGATGGGACAGTGTTTTCACTTGACTTCTGACTCTC
CTACATCAGACATGAAGCAGTCACTTTACAGCATCACAGATAAAATAAAAAATAAAAACATTCTGGAGCT
GTTTGTAGATCTACACATTCTAGTTAAAATATGTAAAATATGAACACAGTTTGCAAATGAGTGAAACAAA
TGATTCCTTTTACTTCTAAAACTCAATGAGATAACAATGAAAAGCAGTGCCTACCTCAGTTGAAAATGGT
TTGCTCATGATGAGCACCACGAAACACAAAATCCATAATTTATTCATGGTTAATGCTGTGTTGTGTTCTT
CCAGTAAGGTTCAGTTCTGGACCGAGGGTGAGTGT
GCATTTAGCAGTTTAATCATGCATACAGACACACTTGCCAGTCAATTACATTAATACACATAATGCCATT
AGAGAAATAGAATCTTTCAAACGCATGATTTAAGTAGTGGAGTTGTTTACCCTGCTAAACTTTATTCACA
TGAAGCAAATAGAAGAAAGGGGCTCTTCCAAGAGAAGTTGCCACTGAGCGTAACCAGACAATCCTTCTTG
GAGGAGTTCTTATTTTCATTAGTTAAAAATCCTTTGAGCTGTGCTGTGCTGTGTTCTGTCTTGCAGATTT
GTGCTCTGTCTGTCAATGACTATCTATTCTGCCACTCCCACATTGATGCTCGCCTTTGGTGAATGTTCAG
GATCTGCTGTGGATGAAGCCCAGGGTGGCAGCCGGGAACAGAAGGTACAGTGTTTTGGGCACAGGACTCA
GTCCTACAGCACTGGAGTTAGTAGATGCGCTGGGCGATGGGACAGTGTTTTCACTTGACTTCTGACTCTC
CTACATCAGACATGAAGCAGTCACTTTACAGCATCACAGATAAAATAAAAAATAAAAACATTCTGGAGCT
GTTTGTAGATCTACACATTCTAGTTAAAATATGTAAAATATGAACACAGTTTGCAAATGAGTGAAACAAA
TGATTCCTTTTACTTCTAAAACTCAATGAGATAACAATGAAAAGCAGTGCCTACCTCAGTTGAAAATGGT
TTGCTCATGATGAGCACCACGAAACACAAAATCCATAATTTATTCATGGTTAATGCTGTGTTGTGTTCTT
CCAGTAAGGTTCAGTTCTGGACCGAGGGTGAGTGT