ZFLNCG09680
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR594416 | retina | transgenic flk1 | 4.53 |
| ERR594417 | retina | normal | 4.07 |
| SRR891495 | heart | normal | 2.96 |
| SRR957181 | heart | normal | 2.93 |
| SRR1648856 | brain | normal | 2.73 |
| SRR1562530 | liver | normal | 2.60 |
| SRR957180 | heart | 7 days after heart tip amputation | 2.57 |
| SRR1291417 | 5 dpi | injected marinum | 2.50 |
| SRR1562528 | eye | normal | 2.10 |
| SRR1291414 | 5 dpi | normal | 1.82 |
Express in tissues
Correlated coding gene
Download
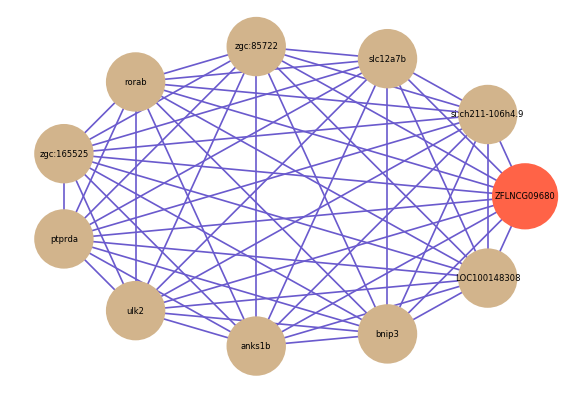
| gene | correlation coefficent |
|---|---|
| si:ch211-106h4.9 | 0.69 |
| slc12a7b | 0.67 |
| zgc:85722 | 0.66 |
| rorab | 0.66 |
| zgc:165525 | 0.66 |
| ptprda | 0.66 |
| ulk2 | 0.65 |
| anks1b | 0.65 |
| bnip3 | 0.64 |
| LOC100148308 | 0.64 |
Gene Ontology
Download
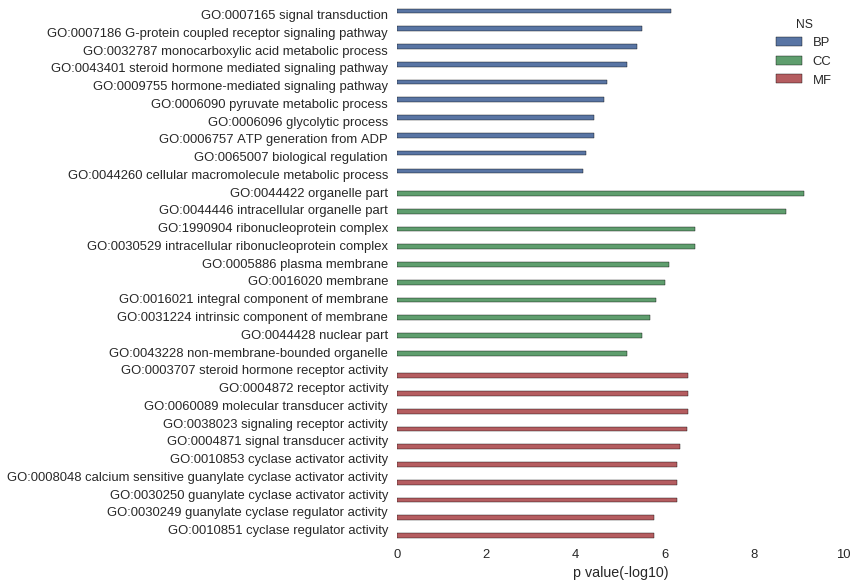
| GO | P value |
|---|---|
| GO:0007165 | 7.38e-07 |
| GO:0007186 | 3.21e-06 |
| GO:0032787 | 4.34e-06 |
| GO:0043401 | 7.28e-06 |
| GO:0009755 | 1.96e-05 |
| GO:0006090 | 2.38e-05 |
| GO:0006096 | 3.97e-05 |
| GO:0006757 | 3.97e-05 |
| GO:0065007 | 5.94e-05 |
| GO:0044260 | 6.93e-05 |
| GO:0044422 | 7.89e-10 |
| GO:0044446 | 1.93e-09 |
| GO:1990904 | 2.13e-07 |
| GO:0030529 | 2.13e-07 |
| GO:0005886 | 8.30e-07 |
| GO:0016020 | 1.01e-06 |
| GO:0016021 | 1.56e-06 |
| GO:0031224 | 2.13e-06 |
| GO:0044428 | 3.25e-06 |
| GO:0043228 | 7.17e-06 |
| GO:0003707 | 3.01e-07 |
| GO:0004872 | 3.11e-07 |
| GO:0060089 | 3.11e-07 |
| GO:0038023 | 3.16e-07 |
| GO:0004871 | 4.59e-07 |
| GO:0010853 | 5.35e-07 |
| GO:0008048 | 5.35e-07 |
| GO:0030250 | 5.35e-07 |
| GO:0030249 | 1.81e-06 |
| GO:0010851 | 1.81e-06 |
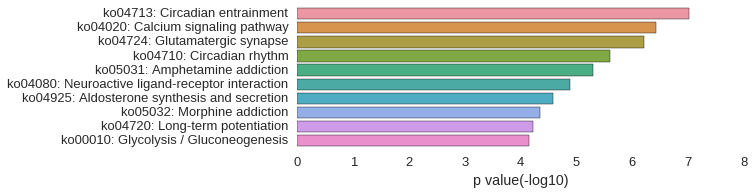
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG09680
ACTTTTAAAAACAAATTCATTTGTCTATCCTCCAAAAACAATCCAATCTTTGAACCAGTTGCTCATTCCA
TAGCGCCCATATGTATTTATTCATCTCTCTGCTGTGTTTTTTCCCCCTTTTACATTCACTGCCGAGGTCT
TAAACTTAACACGTAATTACACAATGGTGAGCCCTGCCTGTGATAAGCTTTTGAATGAATGAGTAGCCGC
TGAGACTAAATTGCTCTCGCTCTCCTGCTCCCTCCTGTCGCGTCTGGCCGTCAGGTCAGGGTTTGAGGCG
AGTTGGCAGTGCGGAGAGGACCAGGCCTGGCCCCGTGCCTCAGTGCCTGCGGGACCCTACCCAGACTGCC
ACGGCCCCTCTGGCCTTCACAATGAATAGAGGTCTTCAGACACCAGTGCACTGAACTGAGGCAGCCTTTT
CAGCAAACAATCCTATCTGCTTCCTCTCCACTCCAGTCCTCCTGCAACTTCACTCAACTCGTCCAGTTTG
TCAGGGGGGCTTATTTTTGTTGTTTTATTTCCATGTGTTCGGGAGGAAAAAAAAAAAAAGAAGAAGAGCT
AGAATGCTGGAAATGGCCCCGAAAAAAACACTGAG
ACTTTTAAAAACAAATTCATTTGTCTATCCTCCAAAAACAATCCAATCTTTGAACCAGTTGCTCATTCCA
TAGCGCCCATATGTATTTATTCATCTCTCTGCTGTGTTTTTTCCCCCTTTTACATTCACTGCCGAGGTCT
TAAACTTAACACGTAATTACACAATGGTGAGCCCTGCCTGTGATAAGCTTTTGAATGAATGAGTAGCCGC
TGAGACTAAATTGCTCTCGCTCTCCTGCTCCCTCCTGTCGCGTCTGGCCGTCAGGTCAGGGTTTGAGGCG
AGTTGGCAGTGCGGAGAGGACCAGGCCTGGCCCCGTGCCTCAGTGCCTGCGGGACCCTACCCAGACTGCC
ACGGCCCCTCTGGCCTTCACAATGAATAGAGGTCTTCAGACACCAGTGCACTGAACTGAGGCAGCCTTTT
CAGCAAACAATCCTATCTGCTTCCTCTCCACTCCAGTCCTCCTGCAACTTCACTCAACTCGTCCAGTTTG
TCAGGGGGGCTTATTTTTGTTGTTTTATTTCCATGTGTTCGGGAGGAAAAAAAAAAAAAGAAGAAGAGCT
AGAATGCTGGAAATGGCCCCGAAAAAAACACTGAG