ZFLNCG09802
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR023144 | brain | normal | 231.48 |
| ERR023143 | swim bladder | normal | 209.65 |
| ERR023145 | heart | normal | 147.45 |
| SRR592702 | pineal gland | normal | 110.64 |
| SRR592703 | pineal gland | normal | 80.86 |
| SRR592699 | pineal gland | normal | 63.15 |
| SRR592701 | pineal gland | normal | 55.05 |
| SRR1565815 | brain | normal | 44.92 |
| SRR1565805 | brain | normal | 33.24 |
| SRR1565813 | brain | normal | 32.00 |
Express in tissues
Correlated coding gene
Download
Gene Ontology
Download
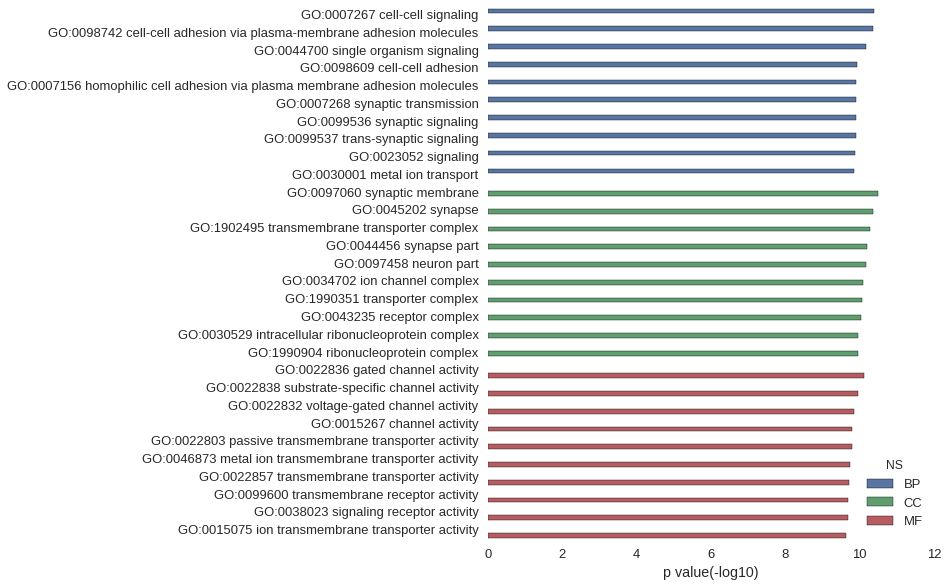
| GO | P value |
|---|---|
| GO:0007267 | 4.15e-11 |
| GO:0098742 | 4.47e-11 |
| GO:0044700 | 6.68e-11 |
| GO:0098609 | 1.20e-10 |
| GO:0007156 | 1.26e-10 |
| GO:0007268 | 1.27e-10 |
| GO:0099536 | 1.27e-10 |
| GO:0099537 | 1.27e-10 |
| GO:0023052 | 1.37e-10 |
| GO:0030001 | 1.45e-10 |
| GO:0097060 | 3.20e-11 |
| GO:0045202 | 4.47e-11 |
| GO:1902495 | 5.18e-11 |
| GO:0044456 | 6.34e-11 |
| GO:0097458 | 6.92e-11 |
| GO:0034702 | 7.99e-11 |
| GO:1990351 | 8.88e-11 |
| GO:0043235 | 9.20e-11 |
| GO:0030529 | 1.12e-10 |
| GO:1990904 | 1.12e-10 |
| GO:0022836 | 7.58e-11 |
| GO:0022838 | 1.08e-10 |
| GO:0022832 | 1.46e-10 |
| GO:0015267 | 1.65e-10 |
| GO:0022803 | 1.65e-10 |
| GO:0046873 | 1.87e-10 |
| GO:0022857 | 1.90e-10 |
| GO:0099600 | 2.02e-10 |
| GO:0038023 | 2.06e-10 |
| GO:0015075 | 2.30e-10 |
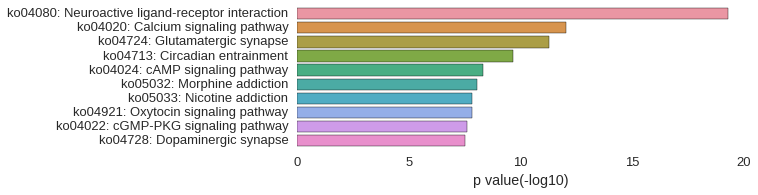
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG09802
TTTTTGCAGTGTAGCTCTTGGATGGAGATAACAGCACTCACACTGAGAGATACAGAGAGTAATATGAAAC
CTGAAGTCTCGAAATGCACTGACTCAATGAAATAAATTAAGTTTATAGTTCGGTTTATGTCTTTTCAAGG
TAAATAAGCACAGAAGATTGACATTGTGCACTATTGAAGACGCACTGTGATACTTTTTTCTTCAGTATTG
TTTAGCACCTTCTCTTTAATGGCATTTTTTTAAGGACCCACACTGCAAACATGATTTTCTTACTTAGATT
TTGTCTTGTTTCTAGTCCAAATACCTAAAAATTCTTTCATAAAGAAGCACTTTCAAG
TTTTTGCAGTGTAGCTCTTGGATGGAGATAACAGCACTCACACTGAGAGATACAGAGAGTAATATGAAAC
CTGAAGTCTCGAAATGCACTGACTCAATGAAATAAATTAAGTTTATAGTTCGGTTTATGTCTTTTCAAGG
TAAATAAGCACAGAAGATTGACATTGTGCACTATTGAAGACGCACTGTGATACTTTTTTCTTCAGTATTG
TTTAGCACCTTCTCTTTAATGGCATTTTTTTAAGGACCCACACTGCAAACATGATTTTCTTACTTAGATT
TTGTCTTGTTTCTAGTCCAAATACCTAAAAATTCTTTCATAAAGAAGCACTTTCAAG