ZFLNCG09849
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR592701 | pineal gland | normal | 27.16 |
| SRR592703 | pineal gland | normal | 23.56 |
| SRR592702 | pineal gland | normal | 20.91 |
| SRR592700 | pineal gland | normal | 15.20 |
| SRR592699 | pineal gland | normal | 13.83 |
| SRR891511 | brain | normal | 12.52 |
| SRR519732 | 7 dpf | FETOH treatment | 12.22 |
| SRR519752 | 7 dpf | VD3 treatment | 11.40 |
| SRR519727 | 6 dpf | FETOH treatment | 11.29 |
| SRR519747 | 6 dpf | VD3 treatment | 10.65 |
Express in tissues
Correlated coding gene
Download
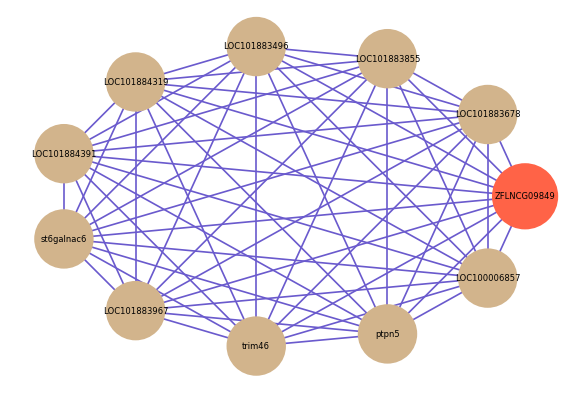
| gene | correlation coefficent |
|---|---|
| LOC101883678 | 0.88 |
| LOC101883855 | 0.84 |
| LOC101883496 | 0.83 |
| LOC101884319 | 0.80 |
| LOC101884391 | 0.79 |
| st6galnac6 | 0.79 |
| LOC101883967 | 0.79 |
| trim46 | 0.78 |
| ptpn5 | 0.78 |
| LOC100006857 | 0.78 |
Gene Ontology
Download
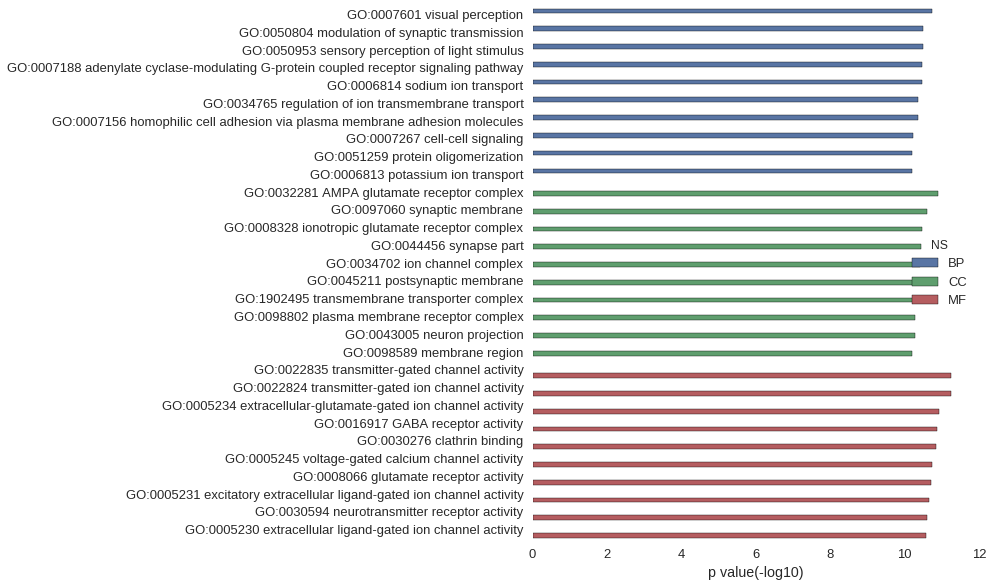
| GO | P value |
|---|---|
| GO:0007601 | 1.80e-11 |
| GO:0050804 | 3.14e-11 |
| GO:0050953 | 3.18e-11 |
| GO:0007188 | 3.45e-11 |
| GO:0006814 | 3.45e-11 |
| GO:0034765 | 4.24e-11 |
| GO:0007156 | 4.29e-11 |
| GO:0007267 | 6.00e-11 |
| GO:0051259 | 6.23e-11 |
| GO:0006813 | 6.24e-11 |
| GO:0032281 | 1.23e-11 |
| GO:0097060 | 2.50e-11 |
| GO:0008328 | 3.40e-11 |
| GO:0044456 | 3.72e-11 |
| GO:0034702 | 3.94e-11 |
| GO:0045211 | 4.39e-11 |
| GO:1902495 | 4.41e-11 |
| GO:0098802 | 5.32e-11 |
| GO:0043005 | 5.32e-11 |
| GO:0098589 | 6.16e-11 |
| GO:0022835 | 5.71e-12 |
| GO:0022824 | 5.71e-12 |
| GO:0005234 | 1.15e-11 |
| GO:0016917 | 1.34e-11 |
| GO:0030276 | 1.43e-11 |
| GO:0005245 | 1.83e-11 |
| GO:0008066 | 1.99e-11 |
| GO:0005231 | 2.14e-11 |
| GO:0030594 | 2.57e-11 |
| GO:0005230 | 2.67e-11 |
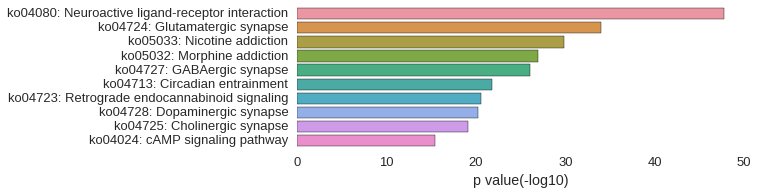
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG09849
ACACACACACACACACACACAGAGACACACACACACACACACACACACACACACAAACACACACACACAC
ACACACACAGTACAGTACAGTATAGTAAACACACACACACATACACACACACACACACACACAGTACAGT
ATAGTAAACACACACACACACACACACACACACAGTACAGTACAGTATAGTAACACACACTCACCCTGGC
GAACTCCACTGGTTTGGGCAGCAGACACATGACCATGACGTCGAAGCGCACGTCACACACCGTCTTCAGC
CACTTGGTGACGAACTGCGGCTTGAGCTTCTCCACCAGGCTGCCCACCTCATCATCCATCATGTAGGCGG
TGGAGTCGAACCACTGGAACACCATGCTGACCAGCCGCGCCAGGCTCTCTGTGGGGGTGTTCTCGCTCGG
GGTGCACAGACTCACCTGAGGAGACGTGTGTGTGTGTGTGTGTTAGTCACTGGCATGGCAACAGCACATA
CAGCAAGTAAACACACACATCTGCATCTACCGAGACCTCCGGGTGCTCCGCTGTCCCCCACAGCCCAAAC
ACATGCGCTATAGAGGGACTGATCA
ACACACACACACACACACACAGAGACACACACACACACACACACACACACACACAAACACACACACACAC
ACACACACAGTACAGTACAGTATAGTAAACACACACACACATACACACACACACACACACACAGTACAGT
ATAGTAAACACACACACACACACACACACACACAGTACAGTACAGTATAGTAACACACACTCACCCTGGC
GAACTCCACTGGTTTGGGCAGCAGACACATGACCATGACGTCGAAGCGCACGTCACACACCGTCTTCAGC
CACTTGGTGACGAACTGCGGCTTGAGCTTCTCCACCAGGCTGCCCACCTCATCATCCATCATGTAGGCGG
TGGAGTCGAACCACTGGAACACCATGCTGACCAGCCGCGCCAGGCTCTCTGTGGGGGTGTTCTCGCTCGG
GGTGCACAGACTCACCTGAGGAGACGTGTGTGTGTGTGTGTGTTAGTCACTGGCATGGCAACAGCACATA
CAGCAAGTAAACACACACATCTGCATCTACCGAGACCTCCGGGTGCTCCGCTGTCCCCCACAGCCCAAAC
ACATGCGCTATAGAGGGACTGATCA