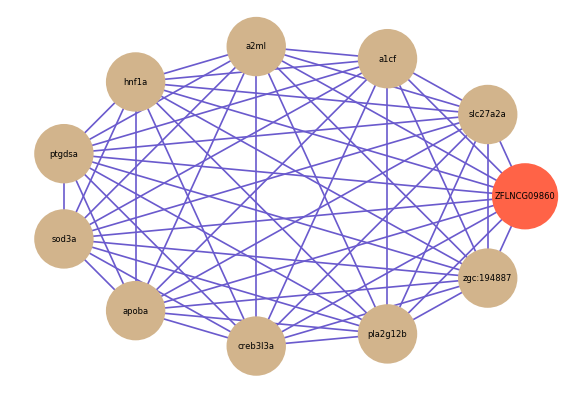
ZFLNCG09860
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1035239 | liver | transgenic mCherry control | 31.06 |
| SRR1562530 | liver | normal | 24.40 |
| SRR797914 | embryo | pbx2/4-MO | 21.47 |
| SRR797917 | embryo | pbx2/4-MO and prdm1-/- | 20.46 |
| SRR797911 | embryo | prdm1-/- | 18.15 |
| SRR797908 | embryo | normal | 17.57 |
| SRR658541 | 6 somite | normal | 16.81 |
| SRR658545 | 6 somite | Gata6 morphant | 14.05 |
| SRR658543 | 6 somite | Gata5 morphan | 13.59 |
| SRR1562529 | intestine and pancreas | normal | 12.34 |
Express in tissues
Correlated coding gene
Download
Gene Ontology
Download
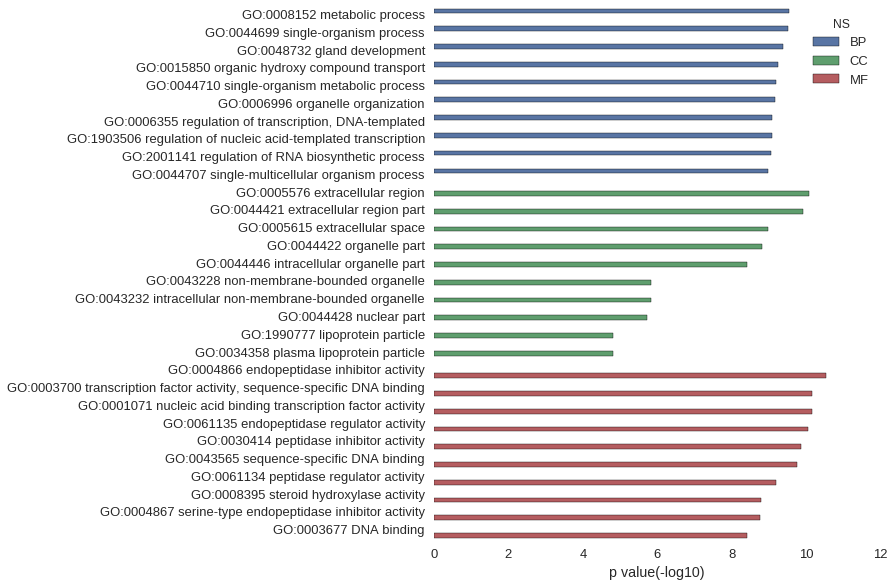
| GO | P value |
|---|---|
| GO:0008152 | 2.86e-10 |
| GO:0044699 | 3.11e-10 |
| GO:0048732 | 4.29e-10 |
| GO:0015850 | 5.52e-10 |
| GO:0044710 | 6.27e-10 |
| GO:0006996 | 6.98e-10 |
| GO:0006355 | 8.13e-10 |
| GO:1903506 | 8.26e-10 |
| GO:2001141 | 8.56e-10 |
| GO:0044707 | 1.05e-09 |
| GO:0005576 | 8.16e-11 |
| GO:0044421 | 1.21e-10 |
| GO:0005615 | 1.03e-09 |
| GO:0044422 | 1.51e-09 |
| GO:0044446 | 3.77e-09 |
| GO:0043228 | 1.43e-06 |
| GO:0043232 | 1.43e-06 |
| GO:0044428 | 1.92e-06 |
| GO:1990777 | 1.54e-05 |
| GO:0034358 | 1.54e-05 |
| GO:0004866 | 2.90e-11 |
| GO:0003700 | 7.10e-11 |
| GO:0001071 | 7.10e-11 |
| GO:0061135 | 8.85e-11 |
| GO:0030414 | 1.35e-10 |
| GO:0043565 | 1.80e-10 |
| GO:0061134 | 6.57e-10 |
| GO:0008395 | 1.62e-09 |
| GO:0004867 | 1.77e-09 |
| GO:0003677 | 3.88e-09 |
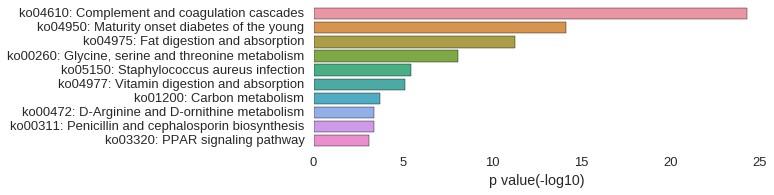
KEGG Pathway
Download
Conservation
| Zebrafish lncRNA Transcript | Human lncRNA Transcript | Mouse lncRNA Transcript | Methods |
|---|---|---|---|
| ZFLNCT15180 | NONMMUT084587; | Direct blastn |
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG09860
CAGATTGATATTCTGAGTTAAATGAGCTCAATCTTAAGTCTGTGTGACTGTCTGGTGGAAAATTATGAGG
TCAAAGTTTGGTCTGATGTTGAGTCTTTGAGACTTTTCTGAGTTTTCCAGGTCAGCTGTCAGGAAAACTG
TAAGTCCGATCAGTGGGTAAAGATAGAGCAACCCGACTCAGAGCAGATTGGAGGTCTGGAGTTAGTCTGG
TGTTTGAAGTATGAGCAGTCTAGGAGATGAGTATAGAGACTAGTCTCAGAAGAAGACGGGAGAATCATCA
GTGGATGTAGGATAACAGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGGACACTAATATGCT
TATATTTATGTATTGATCGTTGTTGTGTTTGCTGTGAACTCTGACCTGGAACTGTAACGCCGTACACATT
TGCAGCCTCAATGCAGTCCAGCATGAGCAGGATCTCCGACACCTCAGTGGTGGCCACGTTCTCTCCCTTC
CACCTGATCAAGCAGAAGCAGACTCATGATGAGCTTCAGCTGCTATTCATGTGTTCTCAGCCATGATGCT
GCTCACACAGCGAGTCACAGACACTGATCATGAGACGGTGAGAACACACAGTGTTCAGTGAGGTGAGAGC
TGCCTCTGTCCAGCGCTTCACTACAGTGAGTCTGTAATCTGGACTCTGCTGTTGGGTCTCTCAGTGATGT
GTGTTGGATGTTTACAGAATGGATCAGCTAATACACACTCTGACGCTCGGTGTGATCGGCGGCGTCTCAC
CGAAACGTGTCTCCGATGCGATCCTGAAAGTAGAGGAATCCCTGGCGGTCGGCCAGGATCAGATCTCCGG
TGTTGAAGTACATGTCTCCTCTCTGAAAGACGTCCCTCAGCCTCTTCTTCTCAGTCTGCGTGGAGTTTTT
AGCGTAGCCTTCGAATGGAGCTAATTTGTGGATCTTCGCCACCAGCAGCCCAGTCTCACCTGTACACATG
TACAGAGTCATTCAGCAGACGCTTCTGTCCAGAGACACGCTCAGCAACTCAACACTGAAGAAGAGCACCA
GCAGAGCTGATGACAGAGGAGCCGATGGTGCGCAGAGCAGAGTGAGATG
CAGATTGATATTCTGAGTTAAATGAGCTCAATCTTAAGTCTGTGTGACTGTCTGGTGGAAAATTATGAGG
TCAAAGTTTGGTCTGATGTTGAGTCTTTGAGACTTTTCTGAGTTTTCCAGGTCAGCTGTCAGGAAAACTG
TAAGTCCGATCAGTGGGTAAAGATAGAGCAACCCGACTCAGAGCAGATTGGAGGTCTGGAGTTAGTCTGG
TGTTTGAAGTATGAGCAGTCTAGGAGATGAGTATAGAGACTAGTCTCAGAAGAAGACGGGAGAATCATCA
GTGGATGTAGGATAACAGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGGACACTAATATGCT
TATATTTATGTATTGATCGTTGTTGTGTTTGCTGTGAACTCTGACCTGGAACTGTAACGCCGTACACATT
TGCAGCCTCAATGCAGTCCAGCATGAGCAGGATCTCCGACACCTCAGTGGTGGCCACGTTCTCTCCCTTC
CACCTGATCAAGCAGAAGCAGACTCATGATGAGCTTCAGCTGCTATTCATGTGTTCTCAGCCATGATGCT
GCTCACACAGCGAGTCACAGACACTGATCATGAGACGGTGAGAACACACAGTGTTCAGTGAGGTGAGAGC
TGCCTCTGTCCAGCGCTTCACTACAGTGAGTCTGTAATCTGGACTCTGCTGTTGGGTCTCTCAGTGATGT
GTGTTGGATGTTTACAGAATGGATCAGCTAATACACACTCTGACGCTCGGTGTGATCGGCGGCGTCTCAC
CGAAACGTGTCTCCGATGCGATCCTGAAAGTAGAGGAATCCCTGGCGGTCGGCCAGGATCAGATCTCCGG
TGTTGAAGTACATGTCTCCTCTCTGAAAGACGTCCCTCAGCCTCTTCTTCTCAGTCTGCGTGGAGTTTTT
AGCGTAGCCTTCGAATGGAGCTAATTTGTGGATCTTCGCCACCAGCAGCCCAGTCTCACCTGTACACATG
TACAGAGTCATTCAGCAGACGCTTCTGTCCAGAGACACGCTCAGCAACTCAACACTGAAGAAGAGCACCA
GCAGAGCTGATGACAGAGGAGCCGATGGTGCGCAGAGCAGAGTGAGATG