ZFLNCG09865
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR592700 | pineal gland | normal | 7.39 |
| SRR891511 | brain | normal | 7.04 |
| SRR1565819 | brain | normal | 4.78 |
| SRR1167757 | embryo | mpeg1 morpholino and bacterial infection | 4.47 |
| ERR023144 | brain | normal | 4.45 |
| SRR1565815 | brain | normal | 4.37 |
| SRR1565817 | brain | normal | 4.19 |
| SRR1565813 | brain | normal | 3.93 |
| SRR1565805 | brain | normal | 3.88 |
| SRR1565807 | brain | normal | 3.80 |
Express in tissues
Correlated coding gene
Download
Gene Ontology
Download
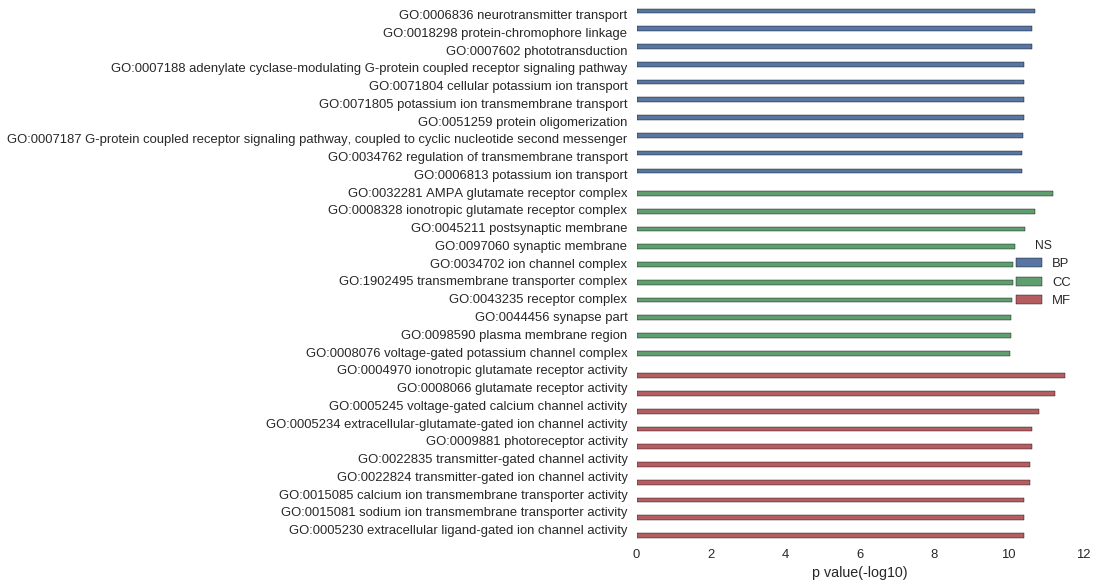
| GO | P value |
|---|---|
| GO:0006836 | 1.93e-11 |
| GO:0018298 | 2.40e-11 |
| GO:0007602 | 2.40e-11 |
| GO:0007188 | 3.76e-11 |
| GO:0071804 | 3.91e-11 |
| GO:0071805 | 3.91e-11 |
| GO:0051259 | 3.92e-11 |
| GO:0007187 | 3.98e-11 |
| GO:0034762 | 4.28e-11 |
| GO:0006813 | 4.28e-11 |
| GO:0032281 | 6.29e-12 |
| GO:0008328 | 1.97e-11 |
| GO:0045211 | 3.52e-11 |
| GO:0097060 | 6.78e-11 |
| GO:0034702 | 7.71e-11 |
| GO:1902495 | 7.76e-11 |
| GO:0043235 | 8.11e-11 |
| GO:0044456 | 8.40e-11 |
| GO:0098590 | 8.47e-11 |
| GO:0008076 | 9.25e-11 |
| GO:0004970 | 3.01e-12 |
| GO:0008066 | 5.77e-12 |
| GO:0005245 | 1.54e-11 |
| GO:0005234 | 2.37e-11 |
| GO:0009881 | 2.40e-11 |
| GO:0022835 | 2.70e-11 |
| GO:0022824 | 2.70e-11 |
| GO:0015085 | 3.79e-11 |
| GO:0015081 | 3.87e-11 |
| GO:0005230 | 3.91e-11 |
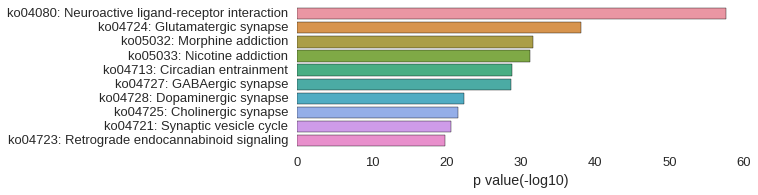
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG09865
CGGCAGCGAATGCTTTGAGATTGAGTGAGTGTTTGAAGAGCTCAGACCTGCAGAACTCACAGGGCATCAG
ACTCCATGATGTCAGGGTTTGTGACTCGTCTGTAGAATCAGAGCTGCCATTCACAAAAAGGCCTGTTTTT
AATAGTGTCAGATAGCTAGCAGACACTCCTGTAGCTTAGCTTCGTCCAGCGTGTTCTTAGTGTAACCTGA
GTAGATCATAGCACAAGGTGAGGGGCGGGGTTTAACCTAGTGGGTGGAGTTATTTGTTAAAAGTGGGCGG
GTTATTTAACAAGTGGGTGGGATATTTGAAGGATTGGCGGGGTTATTTGACATAAGTGGGCGGGGTGTTT
GAGTGAGGAACAAGGTTATTTGACAGGTGGGTGGGGTGTTTGAGTGAAGGGCGGGGTTATTTGACAAGCA
GGTGGGGTGTTCGGTTTAGGGGCAGGGTTATTTGATATAAGTGGGTGGAGTATTTAAGGGAGA
CGGCAGCGAATGCTTTGAGATTGAGTGAGTGTTTGAAGAGCTCAGACCTGCAGAACTCACAGGGCATCAG
ACTCCATGATGTCAGGGTTTGTGACTCGTCTGTAGAATCAGAGCTGCCATTCACAAAAAGGCCTGTTTTT
AATAGTGTCAGATAGCTAGCAGACACTCCTGTAGCTTAGCTTCGTCCAGCGTGTTCTTAGTGTAACCTGA
GTAGATCATAGCACAAGGTGAGGGGCGGGGTTTAACCTAGTGGGTGGAGTTATTTGTTAAAAGTGGGCGG
GTTATTTAACAAGTGGGTGGGATATTTGAAGGATTGGCGGGGTTATTTGACATAAGTGGGCGGGGTGTTT
GAGTGAGGAACAAGGTTATTTGACAGGTGGGTGGGGTGTTTGAGTGAAGGGCGGGGTTATTTGACAAGCA
GGTGGGGTGTTCGGTTTAGGGGCAGGGTTATTTGATATAAGTGGGTGGAGTATTTAAGGGAGA