ZFLNCG10849
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR372787 | 2-4 cell | normal | 34.61 |
| SRR372793 | shield | normal | 22.50 |
| SRR748490 | 1 kcell | normal | 19.16 |
| SRR1049814 | 256 cell | normal | 16.85 |
| SRR372791 | dome | normal | 16.50 |
| SRR1621206 | embryo | normal | 15.53 |
| SRR748489 | egg | normal | 14.75 |
| SRR1021213 | sphere stage | normal | 12.28 |
| SRR748491 | germ ring | normal | 11.40 |
| SRR1021215 | sphere stage | Mzeomesa | 11.29 |
Express in tissues
Correlated coding gene
Download
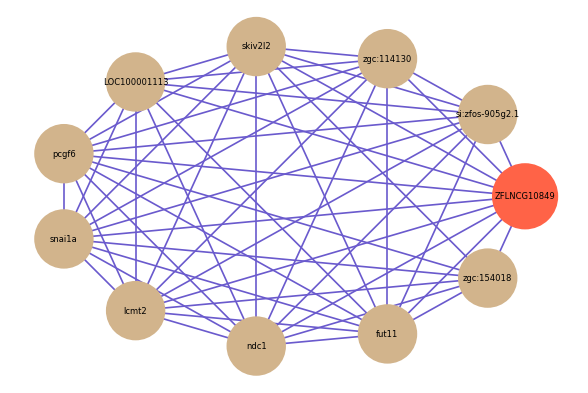
| gene | correlation coefficent |
|---|---|
| si:zfos-905g2.1 | 0.68 |
| zgc:154018 | 0.66 |
| zgc:114130 | 0.66 |
| skiv2l2 | 0.65 |
| LOC100001113 | 0.65 |
| pcgf6 | 0.65 |
| snai1a | 0.65 |
| lcmt2 | 0.65 |
| ndc1 | 0.65 |
| fut11 | 0.65 |
Gene Ontology
Download
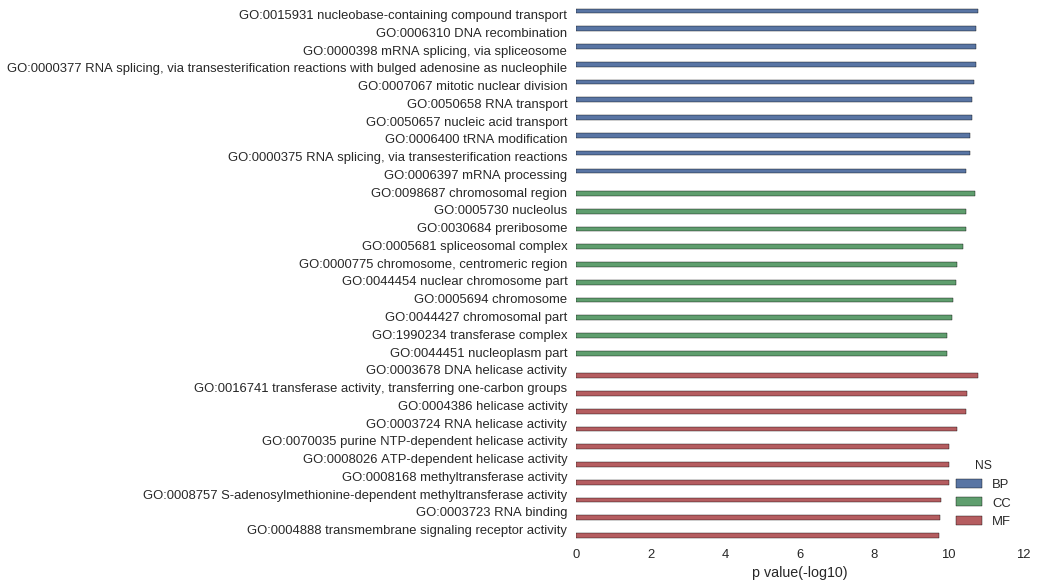
| GO | P value |
|---|---|
| GO:0015931 | 1.56e-11 |
| GO:0006310 | 1.80e-11 |
| GO:0000398 | 1.80e-11 |
| GO:0000377 | 1.80e-11 |
| GO:0007067 | 2.09e-11 |
| GO:0050658 | 2.35e-11 |
| GO:0050657 | 2.35e-11 |
| GO:0006400 | 2.59e-11 |
| GO:0000375 | 2.61e-11 |
| GO:0006397 | 3.32e-11 |
| GO:0098687 | 1.99e-11 |
| GO:0005730 | 3.36e-11 |
| GO:0030684 | 3.39e-11 |
| GO:0005681 | 4.14e-11 |
| GO:0000775 | 5.93e-11 |
| GO:0044454 | 6.44e-11 |
| GO:0005694 | 7.31e-11 |
| GO:0044427 | 7.80e-11 |
| GO:1990234 | 1.06e-10 |
| GO:0044451 | 1.07e-10 |
| GO:0003678 | 1.59e-11 |
| GO:0016741 | 3.14e-11 |
| GO:0004386 | 3.36e-11 |
| GO:0003724 | 5.91e-11 |
| GO:0070035 | 9.52e-11 |
| GO:0008026 | 9.52e-11 |
| GO:0008168 | 9.68e-11 |
| GO:0008757 | 1.54e-10 |
| GO:0003723 | 1.71e-10 |
| GO:0004888 | 1.76e-10 |
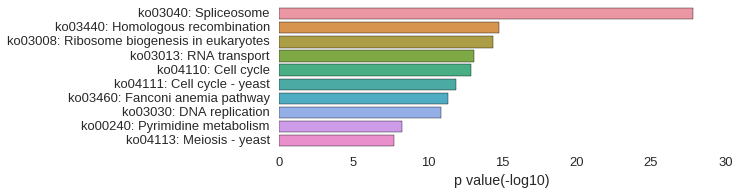
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG10849
ATGAAATGACTTCTCGGTATTAAAGTTAAAATATATTACTTTTTTCCTATCTTGTACTACCTCAAGGGCT
AATGTTTAGGCGAAAATATACAGTAGGCCTAACGTTATTTTGCTTTCAAATACTGCCCCGTCGTGGTTAG
TAACAGAAGTGCAGGGAATACCAAACAAACACACACGGATCTTCAAGCCCTTTCCAAAAAATAAAAAAAT
TTAAAAAAATGCACAGTTAAAACTTCATTAGAATGAAAGAAAAGATTTGATTATAGATGCTTAAAAGCTA
AACACAGAAGACGTGATGAAGAGACCCCAGTGATTAAATCAATTATGCTAATGAAAAGCGTCATTAGAGG
CTTCTACAGGTCAAACTCACCGTCCAAACTGGCTCTTCACAGTTCACAAACACACTCAAACACACTGTAC
AGGAGCTGGGATGTCTGCTAGGGGGATTTTCAGAGGAATAAGATACCTGCAGGTCTTCACGTGCTGGATA
CAAGTAAGTAAATGAAACGAAAAACTGACTATCAACCGTTTCCACTTATTCAGGACTTCGTCAGCTCGCC
AGTCAAAGTTTATAAGTTCCACAAAAAACACCAAAATCGTTAATATTTGTGTAATATTATTCATTTTAAT
TCCCGTTTTAGATGGTGGAATTTATCATTTTCTCCTCTCTGAGGGCTGCAAAGACACGTAAGATGGAGAC
CGAGGCAGACTCAGTAAGTGTTCACTCATGTTGTTGTCTTTTTGTTGCCTCAGACACTAAATGTAAACTG
ATTTTCACCTTTCTAAGGGCATAATTATGACCATGTCAACAAGTTTATATCAAATATTGTAACTAAATCA
AACAAGTGAAAAGCACAA
ATGAAATGACTTCTCGGTATTAAAGTTAAAATATATTACTTTTTTCCTATCTTGTACTACCTCAAGGGCT
AATGTTTAGGCGAAAATATACAGTAGGCCTAACGTTATTTTGCTTTCAAATACTGCCCCGTCGTGGTTAG
TAACAGAAGTGCAGGGAATACCAAACAAACACACACGGATCTTCAAGCCCTTTCCAAAAAATAAAAAAAT
TTAAAAAAATGCACAGTTAAAACTTCATTAGAATGAAAGAAAAGATTTGATTATAGATGCTTAAAAGCTA
AACACAGAAGACGTGATGAAGAGACCCCAGTGATTAAATCAATTATGCTAATGAAAAGCGTCATTAGAGG
CTTCTACAGGTCAAACTCACCGTCCAAACTGGCTCTTCACAGTTCACAAACACACTCAAACACACTGTAC
AGGAGCTGGGATGTCTGCTAGGGGGATTTTCAGAGGAATAAGATACCTGCAGGTCTTCACGTGCTGGATA
CAAGTAAGTAAATGAAACGAAAAACTGACTATCAACCGTTTCCACTTATTCAGGACTTCGTCAGCTCGCC
AGTCAAAGTTTATAAGTTCCACAAAAAACACCAAAATCGTTAATATTTGTGTAATATTATTCATTTTAAT
TCCCGTTTTAGATGGTGGAATTTATCATTTTCTCCTCTCTGAGGGCTGCAAAGACACGTAAGATGGAGAC
CGAGGCAGACTCAGTAAGTGTTCACTCATGTTGTTGTCTTTTTGTTGCCTCAGACACTAAATGTAAACTG
ATTTTCACCTTTCTAAGGGCATAATTATGACCATGTCAACAAGTTTATATCAAATATTGTAACTAAATCA
AACAAGTGAAAAGCACAA