ZFLNCG10869
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR800045 | muscle | normal | 236.59 |
| SRR957180 | heart | 7 days after heart tip amputation | 52.07 |
| SRR957181 | heart | normal | 44.80 |
| ERR023144 | brain | normal | 42.89 |
| ERR023145 | heart | normal | 37.04 |
| SRR065197 | 3 dpf | U1C knockout | 20.13 |
| ERR023143 | swim bladder | normal | 19.62 |
| SRR1565811 | brain | normal | 18.79 |
| SRR726540 | 5 dpf | normal | 17.87 |
| SRR592702 | pineal gland | normal | 17.64 |
Express in tissues
Correlated coding gene
Download
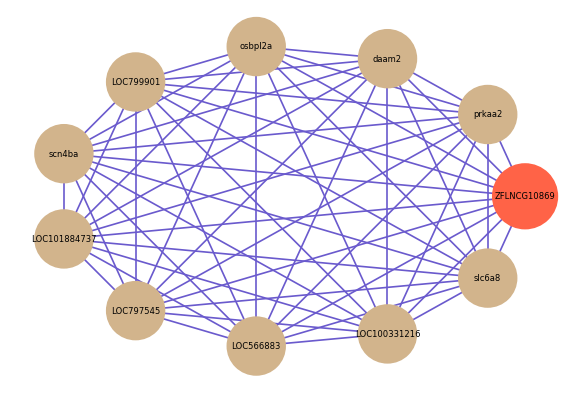
| gene | correlation coefficent |
|---|---|
| prkaa2 | 0.85 |
| daam2 | 0.81 |
| osbpl2a | 0.78 |
| LOC799901 | 0.77 |
| scn4ba | 0.76 |
| LOC101884737 | 0.76 |
| LOC797545 | 0.76 |
| LOC566883 | 0.76 |
| LOC100331216 | 0.75 |
| slc6a8 | 0.75 |
Gene Ontology
Download
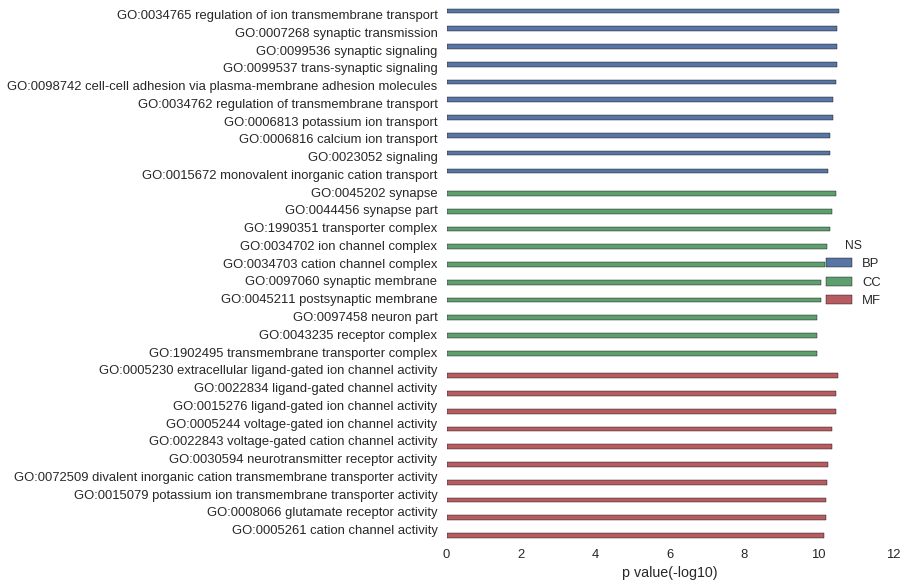
| GO | P value |
|---|---|
| GO:0034765 | 2.87e-11 |
| GO:0007268 | 3.28e-11 |
| GO:0099536 | 3.28e-11 |
| GO:0099537 | 3.28e-11 |
| GO:0098742 | 3.33e-11 |
| GO:0034762 | 4.02e-11 |
| GO:0006813 | 4.02e-11 |
| GO:0006816 | 4.84e-11 |
| GO:0023052 | 4.86e-11 |
| GO:0015672 | 5.54e-11 |
| GO:0045202 | 3.33e-11 |
| GO:0044456 | 4.23e-11 |
| GO:1990351 | 4.88e-11 |
| GO:0034702 | 5.82e-11 |
| GO:0034703 | 6.82e-11 |
| GO:0097060 | 8.60e-11 |
| GO:0045211 | 8.77e-11 |
| GO:0097458 | 1.09e-10 |
| GO:0043235 | 1.11e-10 |
| GO:1902495 | 1.13e-10 |
| GO:0005230 | 2.94e-11 |
| GO:0022834 | 3.38e-11 |
| GO:0015276 | 3.38e-11 |
| GO:0005244 | 4.23e-11 |
| GO:0022843 | 4.30e-11 |
| GO:0030594 | 5.59e-11 |
| GO:0072509 | 5.92e-11 |
| GO:0015079 | 6.12e-11 |
| GO:0008066 | 6.38e-11 |
| GO:0005261 | 7.25e-11 |
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG10869
TTTGGTTTGCAGTAAATCCCTGATGATACACACCATCTTCATCCGGATCAGATGTAAAGTGTGTGTTTCT
GAGGCCTTGCTAATATCACAGCTGCTTTAGTTTAGACAGACAGATATGCTGGACGAGGACAGCCAGAGCC
TAAATGTTGTGCCTTGATATGGTGAAAATGCAGATAGAGACGGATGAGGCACAGAATGTGCCAGTCATTT
TGTTGATATGAGTAGACCTTTAATTAGGCGGCCTTAAAATATTCTCCATGGTTGAGATTATTACAGACAT
ACACGGCCTGGCAGACATTATTAAAGAAATTTCTCTGAACAACACATTCAGC
TTTGGTTTGCAGTAAATCCCTGATGATACACACCATCTTCATCCGGATCAGATGTAAAGTGTGTGTTTCT
GAGGCCTTGCTAATATCACAGCTGCTTTAGTTTAGACAGACAGATATGCTGGACGAGGACAGCCAGAGCC
TAAATGTTGTGCCTTGATATGGTGAAAATGCAGATAGAGACGGATGAGGCACAGAATGTGCCAGTCATTT
TGTTGATATGAGTAGACCTTTAATTAGGCGGCCTTAAAATATTCTCCATGGTTGAGATTATTACAGACAT
ACACGGCCTGGCAGACATTATTAAAGAAATTTCTCTGAACAACACATTCAGC