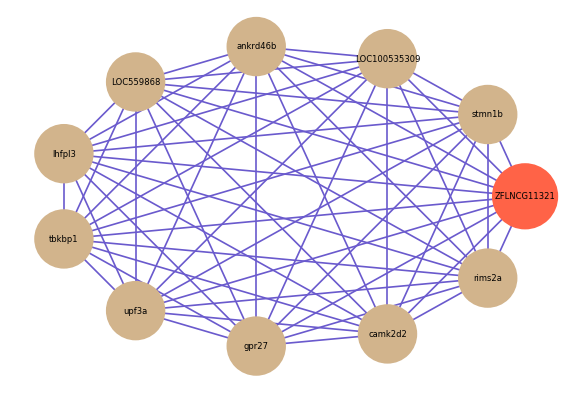
ZFLNCG11321
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1648855 | brain | normal | 64.65 |
| SRR1648854 | brain | normal | 60.99 |
| SRR1648856 | brain | normal | 43.59 |
| ERR023144 | brain | normal | 39.90 |
| SRR592700 | pineal gland | normal | 18.64 |
| SRR592702 | pineal gland | normal | 14.11 |
| SRR942772 | embryo | myd88+/+ and infection with Mycobacterium marinum | 12.63 |
| SRR372802 | 5 dpf | normal | 11.65 |
| SRR1291417 | 5 dpi | injected marinum | 10.71 |
| SRR630463 | 5 dpf | normal | 10.08 |
Express in tissues
Correlated coding gene
Download
Gene Ontology
Download
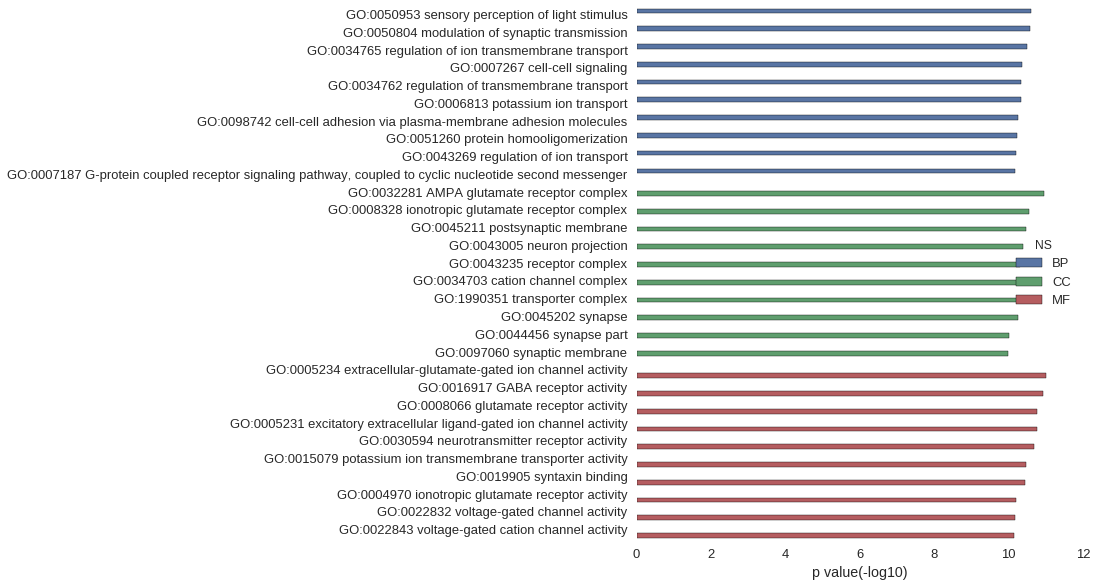
| GO | P value |
|---|---|
| GO:0050953 | 2.52e-11 |
| GO:0050804 | 2.59e-11 |
| GO:0034765 | 3.18e-11 |
| GO:0007267 | 4.22e-11 |
| GO:0034762 | 4.69e-11 |
| GO:0006813 | 4.69e-11 |
| GO:0098742 | 5.73e-11 |
| GO:0051260 | 5.84e-11 |
| GO:0043269 | 6.37e-11 |
| GO:0007187 | 6.56e-11 |
| GO:0032281 | 1.09e-11 |
| GO:0008328 | 2.88e-11 |
| GO:0045211 | 3.47e-11 |
| GO:0043005 | 4.14e-11 |
| GO:0043235 | 4.83e-11 |
| GO:0034703 | 5.54e-11 |
| GO:1990351 | 5.70e-11 |
| GO:0045202 | 5.73e-11 |
| GO:0044456 | 9.98e-11 |
| GO:0097060 | 1.03e-10 |
| GO:0005234 | 9.93e-12 |
| GO:0016917 | 1.17e-11 |
| GO:0008066 | 1.69e-11 |
| GO:0005231 | 1.71e-11 |
| GO:0030594 | 2.03e-11 |
| GO:0015079 | 3.35e-11 |
| GO:0019905 | 3.69e-11 |
| GO:0004970 | 6.13e-11 |
| GO:0022832 | 6.79e-11 |
| GO:0022843 | 6.95e-11 |
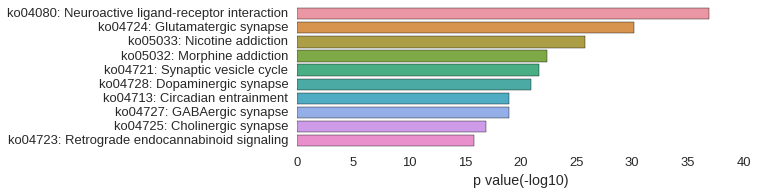
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG11321
TAAATTATAGCATGTCACCCTAAAGGCGTGATTTACCGAATCATTCAATTGGCTCATTCAAAAGGGCTGA
TTCATTTCAAAACCATTTTGTTTTATTTCTCAGTAAACCAGATTTTATACTAGTTATGTATAAAGACTTG
TGTTTTTAAATGAATCAATCATTCTGAATGAATTTATGGAATGAATTACTCACTGAATCTCCCATTAACC
CTTTATCAGGCAATGGATAATTCATGCACAGTGTTGCTCTGAGATGCCCCAATTAGAATTTTTTTATTTT
ATTATTAATCAAAAAATTGTCTGTGCAGATGGCCTAGTGGTTGGTGCGTCGAC
TAAATTATAGCATGTCACCCTAAAGGCGTGATTTACCGAATCATTCAATTGGCTCATTCAAAAGGGCTGA
TTCATTTCAAAACCATTTTGTTTTATTTCTCAGTAAACCAGATTTTATACTAGTTATGTATAAAGACTTG
TGTTTTTAAATGAATCAATCATTCTGAATGAATTTATGGAATGAATTACTCACTGAATCTCCCATTAACC
CTTTATCAGGCAATGGATAATTCATGCACAGTGTTGCTCTGAGATGCCCCAATTAGAATTTTTTTATTTT
ATTATTAATCAAAAAATTGTCTGTGCAGATGGCCTAGTGGTTGGTGCGTCGAC