ZFLNCG11420
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR023144 | brain | normal | 47.93 |
| ERR023145 | heart | normal | 45.49 |
| ERR023146 | head kidney | normal | 26.12 |
| ERR023143 | swim bladder | normal | 24.41 |
| SRR1562530 | liver | normal | 24.30 |
| ERR145635 | skeletal muscle | 27 degree_C to 16 degree_C | 14.74 |
| SRR1565809 | brain | normal | 13.99 |
| SRR1562529 | intestine and pancreas | normal | 13.16 |
| ERR145631 | skeletal muscle | 32 degree_C to 16 degree_C | 13.07 |
| SRR1565811 | brain | normal | 12.93 |
Express in tissues
Correlated coding gene
Download
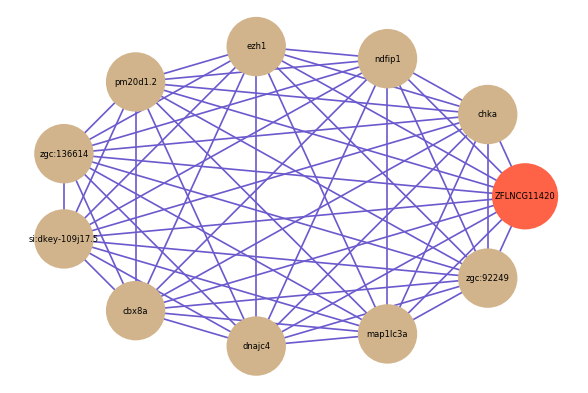
| gene | correlation coefficent |
|---|---|
| chka | 0.81 |
| ndfip1 | 0.80 |
| ezh1 | 0.80 |
| pm20d1.2 | 0.79 |
| zgc:136614 | 0.79 |
| si:dkey-109j17.5 | 0.79 |
| cbx8a | 0.78 |
| dnajc4 | 0.78 |
| map1lc3a | 0.78 |
| zgc:92249 | 0.78 |
Gene Ontology
Download
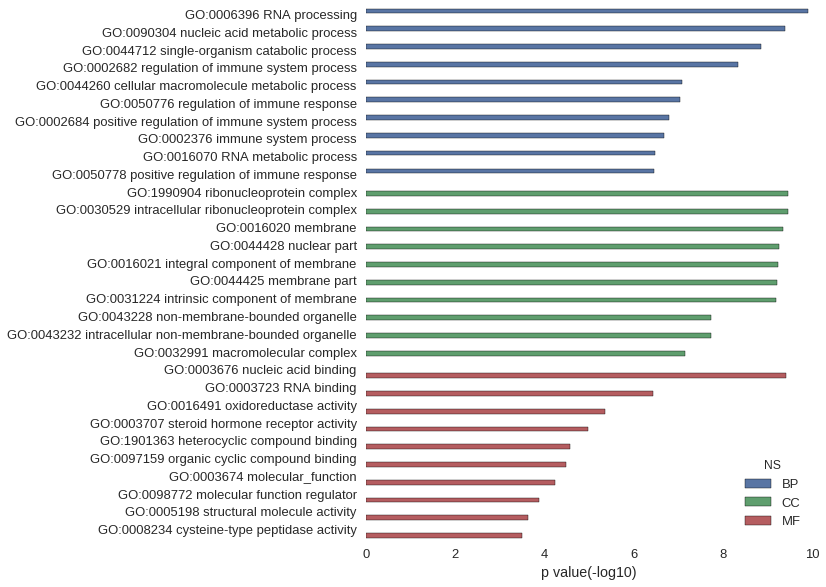
| GO | P value |
|---|---|
| GO:0006396 | 1.24e-10 |
| GO:0090304 | 4.10e-10 |
| GO:0044712 | 1.41e-09 |
| GO:0002682 | 4.65e-09 |
| GO:0044260 | 8.13e-08 |
| GO:0050776 | 9.20e-08 |
| GO:0002684 | 1.65e-07 |
| GO:0002376 | 2.11e-07 |
| GO:0016070 | 3.42e-07 |
| GO:0050778 | 3.47e-07 |
| GO:1990904 | 3.44e-10 |
| GO:0030529 | 3.44e-10 |
| GO:0016020 | 4.57e-10 |
| GO:0044428 | 5.46e-10 |
| GO:0016021 | 5.88e-10 |
| GO:0044425 | 6.20e-10 |
| GO:0031224 | 6.40e-10 |
| GO:0043228 | 1.83e-08 |
| GO:0043232 | 1.83e-08 |
| GO:0032991 | 7.22e-08 |
| GO:0003676 | 3.82e-10 |
| GO:0003723 | 3.80e-07 |
| GO:0016491 | 4.32e-06 |
| GO:0003707 | 1.04e-05 |
| GO:1901363 | 2.68e-05 |
| GO:0097159 | 3.30e-05 |
| GO:0003674 | 5.72e-05 |
| GO:0098772 | 1.35e-04 |
| GO:0005198 | 2.32e-04 |
| GO:0008234 | 3.12e-04 |
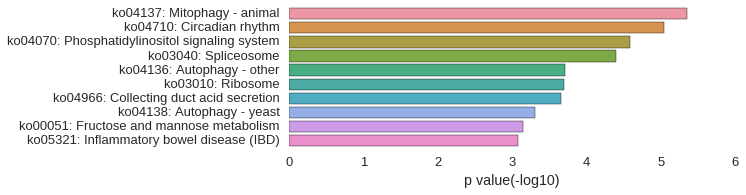
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG11420
ATTAAATTGTGCAAATTAAAATTGTGAACTGAAAATACGCCTTATGGAAACGTGTCCATTTCACAAAAAC
ACCGACGTAGTGCAAAAAAATTTTTTTACACACAGAATTGAAATATATCGCAAACTGCAATGGAAATGTT
TTCTTCCCCATATTAACAGGTCTTGGGAATACGTCGAGAAGTCGTAAGATCATATTTTAAAGACAGTTTG
GTGTGTATATAATGATGTATAATGTATCTGTAAGAAGGCGAAGATTACTATGAACGTATTTTATGCTAAA
TTTTGAACGTCATTATTGAGTAAATCATGACAGATATTGCAGAAAGTTTGGTGGAAATGTAATTTTGCCA
TTCATTTGCGCAAATTAAAACTGCGAACTGAAAATACACCTTATGGAAACGTGTCCATTTCACGAAGGCA
CCTACATAGCGCAAAAGTTTCATTTTTTATGCGCACAATTGAAATGTTTTACAAACTGCAATGCAAAAAG
TTTTTCCCACAAGGACAGGTCTTGGGGCATACGTCAAGAAGTCGTAAGATCATTCTCTATAGACGGTTTG
GTATGTATATGGTGATGTATAGGTAAGAAGGCTTTGATTGCAACTGACTATTATTTTATGCGAAATTTTG
AATGTTATTATTATGTAAATGATGACAGATATTGCACGGCGTTAAGTGTAAATGTAGTAATTTGTCCATT
TAAACTAAAAATACACCTTATGGAAACGTGTCCATTTGCCGATAACACCTACATAGTGCAAAAGTTTTTT
TTGTTTGTTTGTTTTTTTCCACACAGAATTTAAATATTTTGGAAACTGCAAAAGAAACGTTTTTTCCTAC
ATTGACAGATCTTGGGACATACGTCGTGAAGTCGTACGATCATTCTGTAAAGACGGTTTGGTATGTATAT
GATGATGTATAACGTATAGGTAACTAGGATGTGATTGCTATGAACTTATTTTCTGCAAAATTGTGAATGT
CGTTTTTATGTTGAGGATGTTGAGAGTAGATGAAGCTGTTTTTCATTCAATTGCGCAAATTAAAATTGCG
AACTGAAAATAAGCCTTATGGAAACGTGTCCGTTTCACAAAAACATCCCACATTAGATTTCTTTTATGCA
CAAAATAGAAATAATTCGCGAACTTCAATGGAAACGTTTTTTTCCCCCCAC
ATTAAATTGTGCAAATTAAAATTGTGAACTGAAAATACGCCTTATGGAAACGTGTCCATTTCACAAAAAC
ACCGACGTAGTGCAAAAAAATTTTTTTACACACAGAATTGAAATATATCGCAAACTGCAATGGAAATGTT
TTCTTCCCCATATTAACAGGTCTTGGGAATACGTCGAGAAGTCGTAAGATCATATTTTAAAGACAGTTTG
GTGTGTATATAATGATGTATAATGTATCTGTAAGAAGGCGAAGATTACTATGAACGTATTTTATGCTAAA
TTTTGAACGTCATTATTGAGTAAATCATGACAGATATTGCAGAAAGTTTGGTGGAAATGTAATTTTGCCA
TTCATTTGCGCAAATTAAAACTGCGAACTGAAAATACACCTTATGGAAACGTGTCCATTTCACGAAGGCA
CCTACATAGCGCAAAAGTTTCATTTTTTATGCGCACAATTGAAATGTTTTACAAACTGCAATGCAAAAAG
TTTTTCCCACAAGGACAGGTCTTGGGGCATACGTCAAGAAGTCGTAAGATCATTCTCTATAGACGGTTTG
GTATGTATATGGTGATGTATAGGTAAGAAGGCTTTGATTGCAACTGACTATTATTTTATGCGAAATTTTG
AATGTTATTATTATGTAAATGATGACAGATATTGCACGGCGTTAAGTGTAAATGTAGTAATTTGTCCATT
TAAACTAAAAATACACCTTATGGAAACGTGTCCATTTGCCGATAACACCTACATAGTGCAAAAGTTTTTT
TTGTTTGTTTGTTTTTTTCCACACAGAATTTAAATATTTTGGAAACTGCAAAAGAAACGTTTTTTCCTAC
ATTGACAGATCTTGGGACATACGTCGTGAAGTCGTACGATCATTCTGTAAAGACGGTTTGGTATGTATAT
GATGATGTATAACGTATAGGTAACTAGGATGTGATTGCTATGAACTTATTTTCTGCAAAATTGTGAATGT
CGTTTTTATGTTGAGGATGTTGAGAGTAGATGAAGCTGTTTTTCATTCAATTGCGCAAATTAAAATTGCG
AACTGAAAATAAGCCTTATGGAAACGTGTCCGTTTCACAAAAACATCCCACATTAGATTTCTTTTATGCA
CAAAATAGAAATAATTCGCGAACTTCAATGGAAACGTTTTTTTCCCCCCAC