ZFLNCG11619
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1562532 | spleen | normal | 1980.20 |
| SRR1039571 | gastrointestinal | diet 0.1 NPM | 1615.03 |
| SRR1562529 | intestine and pancreas | normal | 1320.49 |
| SRR1039573 | gastrointestinal | diet 0.75 NPM | 1128.24 |
| SRR1647683 | spleen | SVCV treatment | 1087.30 |
| SRR1534350 | larvae | 5 ug/l BDE47 treatment | 873.32 |
| SRR1534349 | larvae | DMSO vehicle | 836.75 |
| SRR1035239 | liver | transgenic mCherry control | 728.13 |
| SRR1534351 | larvae | 500 ug/l BDE47 treatment | 630.51 |
| SRR372802 | 5 dpf | normal | 462.76 |
Express in tissues
Correlated coding gene
Download
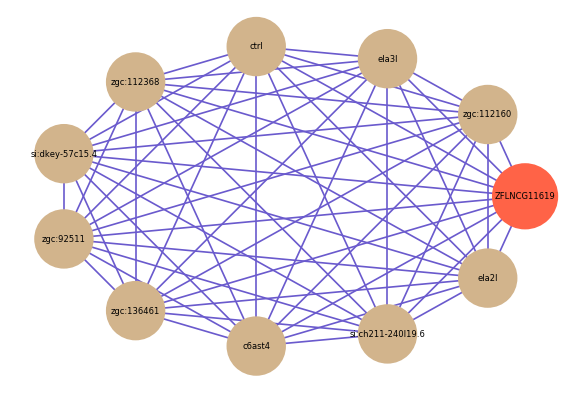
| gene | correlation coefficent |
|---|---|
| zgc:112160 | 0.91 |
| ela3l | 0.90 |
| ctrl | 0.90 |
| zgc:112368 | 0.89 |
| si:dkey-57c15.4 | 0.89 |
| zgc:92511 | 0.88 |
| zgc:136461 | 0.87 |
| c6ast4 | 0.87 |
| si:ch211-240l19.6 | 0.87 |
| ela2l | 0.86 |
Gene Ontology
Download
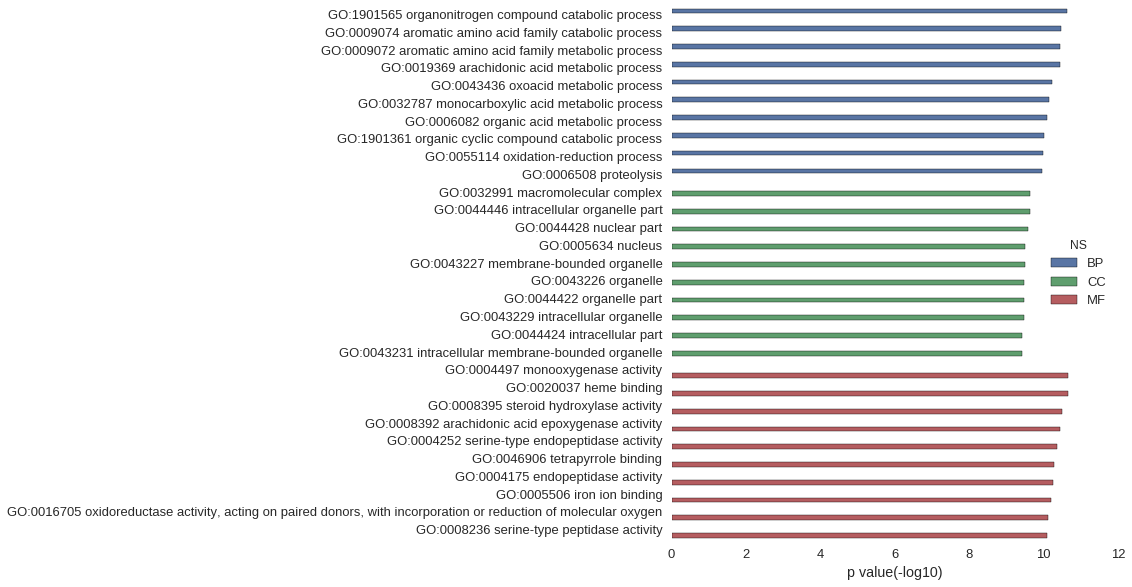
| GO | P value |
|---|---|
| GO:1901565 | 2.40e-11 |
| GO:0009074 | 3.29e-11 |
| GO:0009072 | 3.55e-11 |
| GO:0019369 | 3.56e-11 |
| GO:0043436 | 6.05e-11 |
| GO:0032787 | 6.97e-11 |
| GO:0006082 | 8.04e-11 |
| GO:1901361 | 9.77e-11 |
| GO:0055114 | 1.05e-10 |
| GO:0006508 | 1.07e-10 |
| GO:0032991 | 2.31e-10 |
| GO:0044446 | 2.35e-10 |
| GO:0044428 | 2.56e-10 |
| GO:0005634 | 3.16e-10 |
| GO:0043227 | 3.20e-10 |
| GO:0043226 | 3.25e-10 |
| GO:0044422 | 3.30e-10 |
| GO:0043229 | 3.42e-10 |
| GO:0044424 | 3.74e-10 |
| GO:0043231 | 3.85e-10 |
| GO:0004497 | 2.24e-11 |
| GO:0020037 | 2.24e-11 |
| GO:0008395 | 3.21e-11 |
| GO:0008392 | 3.55e-11 |
| GO:0004252 | 4.22e-11 |
| GO:0046906 | 5.20e-11 |
| GO:0004175 | 5.69e-11 |
| GO:0005506 | 6.49e-11 |
| GO:0016705 | 7.74e-11 |
| GO:0008236 | 8.04e-11 |
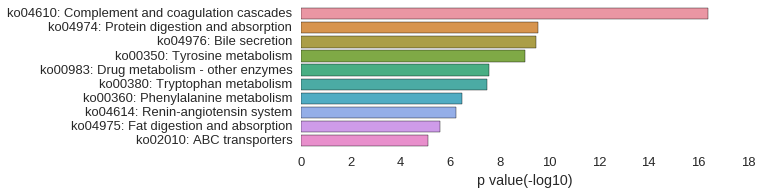
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG11619
CTCACCAAGCTGAAAACATGAGGGGTCTAATTGCTCTTCTCCTGGTGGCTCTGTGCCTCGAGGGCCTCAA
TGCAGCGTGTCCTGATCATGCCACACTGAAAGACGACTATGGCAGCAAACTCTGTGCCCAGATGTTTGAG
CACAGCAGCTACTACTATGACGAGAGCTGCACTGGCAGTTTTCTGTATGTGTATCCTGATGAAGACACCC
CCATCATGCCATGGGTCTGGGAGAACCGAATTTCCTCTCTGCTGGTGGGACGTGGCTGCTCGCTGACGGT
GTGGTCCAAATCCAAGAAGAAGGGAGACAAGAAGAAGTTTTCTGCTGGAGCTGCCTACCACCTGAAGGAG
TATAAGAGAGGCCTGTTCGGAAACTGGAATGACGCCATCTCTGGATACTACTGCACATGCTAGACCTGAA
GACAGAGACTCAAAAGACATTAATGTATAAGGTCAATAATGTAGAAGATCAGCTTTAACCTTGTGTTTTT
GGAAATAATTTTAAATTCAGCTTAATGTAATTCTGCTTAATGACTGAATAAAATATCACAGGTGAAGCGA
AATTATTCCATACCTGGTGCATGATTATTTTTAACTGCATTAAATGCATTGAATAAATCTTATTTTAAAA
GTA
CTCACCAAGCTGAAAACATGAGGGGTCTAATTGCTCTTCTCCTGGTGGCTCTGTGCCTCGAGGGCCTCAA
TGCAGCGTGTCCTGATCATGCCACACTGAAAGACGACTATGGCAGCAAACTCTGTGCCCAGATGTTTGAG
CACAGCAGCTACTACTATGACGAGAGCTGCACTGGCAGTTTTCTGTATGTGTATCCTGATGAAGACACCC
CCATCATGCCATGGGTCTGGGAGAACCGAATTTCCTCTCTGCTGGTGGGACGTGGCTGCTCGCTGACGGT
GTGGTCCAAATCCAAGAAGAAGGGAGACAAGAAGAAGTTTTCTGCTGGAGCTGCCTACCACCTGAAGGAG
TATAAGAGAGGCCTGTTCGGAAACTGGAATGACGCCATCTCTGGATACTACTGCACATGCTAGACCTGAA
GACAGAGACTCAAAAGACATTAATGTATAAGGTCAATAATGTAGAAGATCAGCTTTAACCTTGTGTTTTT
GGAAATAATTTTAAATTCAGCTTAATGTAATTCTGCTTAATGACTGAATAAAATATCACAGGTGAAGCGA
AATTATTCCATACCTGGTGCATGATTATTTTTAACTGCATTAAATGCATTGAATAAATCTTATTTTAAAA
GTA