ZFLNCG11967
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1647684 | spleen | SVCV treatment | 80.91 |
| ERR023145 | heart | normal | 46.42 |
| SRR1647681 | head kidney | SVCV treatment | 41.13 |
| SRR1562529 | intestine and pancreas | normal | 28.92 |
| ERR023143 | swim bladder | normal | 23.31 |
| SRR1647683 | spleen | SVCV treatment | 21.41 |
| ERR023146 | head kidney | normal | 17.21 |
| SRR1562528 | eye | normal | 16.49 |
| SRR1647682 | spleen | normal | 12.10 |
| SRR516121 | skin | male and 3.5 year | 10.08 |
Express in tissues
Correlated coding gene
Download
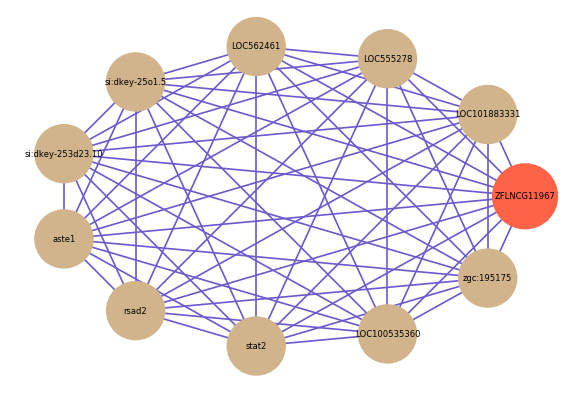
| gene | correlation coefficent |
|---|---|
| LOC101883331 | 0.86 |
| LOC555278 | 0.82 |
| LOC562461 | 0.79 |
| si:dkey-25o1.5 | 0.78 |
| si:dkey-253d23.10 | 0.78 |
| aste1 | 0.77 |
| rsad2 | 0.77 |
| stat2 | 0.77 |
| LOC100535360 | 0.76 |
| zgc:195175 | 0.76 |
Gene Ontology
Download
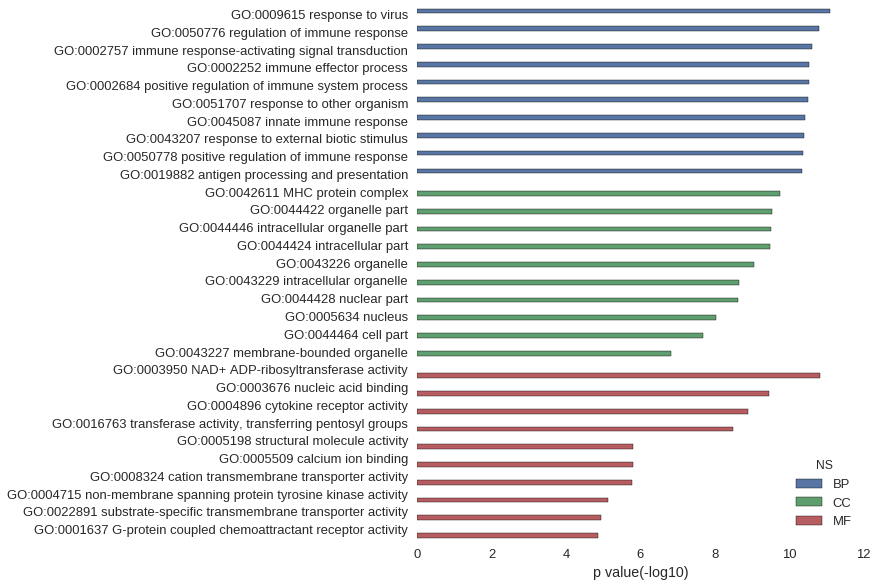
| GO | P value |
|---|---|
| GO:0009615 | 8.15e-12 |
| GO:0050776 | 1.61e-11 |
| GO:0002757 | 2.39e-11 |
| GO:0002252 | 2.90e-11 |
| GO:0002684 | 3.00e-11 |
| GO:0051707 | 3.21e-11 |
| GO:0045087 | 3.73e-11 |
| GO:0043207 | 3.89e-11 |
| GO:0050778 | 4.29e-11 |
| GO:0019882 | 4.62e-11 |
| GO:0042611 | 1.73e-10 |
| GO:0044422 | 2.81e-10 |
| GO:0044446 | 3.01e-10 |
| GO:0044424 | 3.24e-10 |
| GO:0043226 | 8.90e-10 |
| GO:0043229 | 2.24e-09 |
| GO:0044428 | 2.42e-09 |
| GO:0005634 | 9.54e-09 |
| GO:0044464 | 2.01e-08 |
| GO:0043227 | 1.48e-07 |
| GO:0003950 | 1.47e-11 |
| GO:0003676 | 3.53e-10 |
| GO:0004896 | 1.24e-09 |
| GO:0016763 | 3.20e-09 |
| GO:0005198 | 1.55e-06 |
| GO:0005509 | 1.58e-06 |
| GO:0008324 | 1.69e-06 |
| GO:0004715 | 7.56e-06 |
| GO:0022891 | 1.12e-05 |
| GO:0001637 | 1.35e-05 |
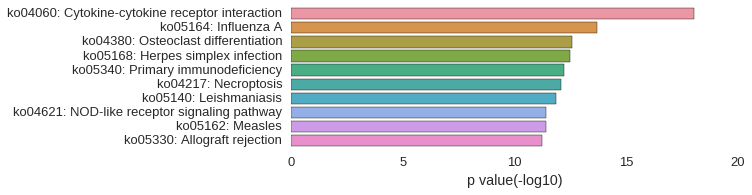
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG11967
ACAATTGCACACATTTAACCAACTTGACAACCTTAACTCAGTTATTTCGAGTTAGTATTAAAACAAGCAT
TTTTACACATATATTTGCTCGGAAATTAATTAAGTGTAAATTTGAACTCAAACAAGTTACAAACTCCAGC
TTTAAGCAGTAATATGCACTATATCGCACACATTTAAAATGTAACGCTCTTCACTATCAACAAAGTTCAT
ACTAGTTTGAACCTGCTCTTCTGTATCAGGACATAACAATAACAAATACTATTTTGCTCTTGGCAGGCCT
TAGAACGTCACAACGTCACATGACCTCTCACACCGTGTGATTATTTATTGCACAAAAATAAAGGTGAGAT
AAAAAAGCCATCTATGATTCAGATCCTGTACATGTACTTTCGATTACATTAGCTTTAGTTATACAGCAGA
TGCTTAAAACTGACGAGGCATTCAGTGATTCAACACGAAGAGGCAATACACACAAAAAGTGCAAGATATG
AAGAATAACTGTGCTCAGAGAATTGGGTGCTAGAGTAAAGGGAGTTTTTTAAAGAGAGACAGAGAAGACT
GTTTCTACAAGTAGTCTCTCAAGCCCACTCACTTCATAAATGCAGTGTTTTTATATGTGTGTGTGAGTGT
CTGTTTATTGTAAAATATACGTGTATTCCTACAATCAACACACACACACACACACACACACACACACACA
CACACACACACACACAAAACCCC
ACAATTGCACACATTTAACCAACTTGACAACCTTAACTCAGTTATTTCGAGTTAGTATTAAAACAAGCAT
TTTTACACATATATTTGCTCGGAAATTAATTAAGTGTAAATTTGAACTCAAACAAGTTACAAACTCCAGC
TTTAAGCAGTAATATGCACTATATCGCACACATTTAAAATGTAACGCTCTTCACTATCAACAAAGTTCAT
ACTAGTTTGAACCTGCTCTTCTGTATCAGGACATAACAATAACAAATACTATTTTGCTCTTGGCAGGCCT
TAGAACGTCACAACGTCACATGACCTCTCACACCGTGTGATTATTTATTGCACAAAAATAAAGGTGAGAT
AAAAAAGCCATCTATGATTCAGATCCTGTACATGTACTTTCGATTACATTAGCTTTAGTTATACAGCAGA
TGCTTAAAACTGACGAGGCATTCAGTGATTCAACACGAAGAGGCAATACACACAAAAAGTGCAAGATATG
AAGAATAACTGTGCTCAGAGAATTGGGTGCTAGAGTAAAGGGAGTTTTTTAAAGAGAGACAGAGAAGACT
GTTTCTACAAGTAGTCTCTCAAGCCCACTCACTTCATAAATGCAGTGTTTTTATATGTGTGTGTGAGTGT
CTGTTTATTGTAAAATATACGTGTATTCCTACAATCAACACACACACACACACACACACACACACACACA
CACACACACACACACAAAACCCC