ZFLNCG12488
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1648856 | brain | normal | 125.13 |
| SRR372802 | 5 dpf | normal | 95.61 |
| ERR023144 | brain | normal | 76.89 |
| ERR023146 | head kidney | normal | 53.35 |
| SRR1648854 | brain | normal | 47.47 |
| SRR1562529 | intestine and pancreas | normal | 42.97 |
| SRR700534 | heart | control morpholino | 32.92 |
| SRR1188148 | embryo | Control PBS 0.5 hpi | 30.89 |
| SRR1205172 | 5dpf | transgenic sqET20 and neomycin treated 5h | 30.81 |
| SRR1291417 | 5 dpi | injected marinum | 30.69 |
Express in tissues
Correlated coding gene
Download
Gene Ontology
Download
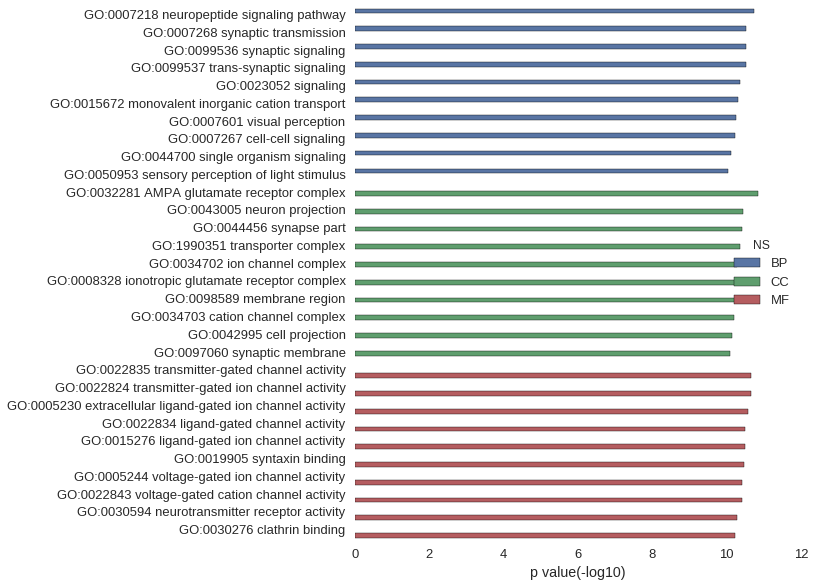
| GO | P value |
|---|---|
| GO:0007218 | 1.79e-11 |
| GO:0007268 | 3.03e-11 |
| GO:0099536 | 3.03e-11 |
| GO:0099537 | 3.03e-11 |
| GO:0023052 | 4.41e-11 |
| GO:0015672 | 5.01e-11 |
| GO:0007601 | 5.58e-11 |
| GO:0007267 | 6.16e-11 |
| GO:0044700 | 7.89e-11 |
| GO:0050953 | 8.97e-11 |
| GO:0032281 | 1.46e-11 |
| GO:0043005 | 3.63e-11 |
| GO:0044456 | 3.88e-11 |
| GO:1990351 | 4.46e-11 |
| GO:0034702 | 5.35e-11 |
| GO:0008328 | 5.91e-11 |
| GO:0098589 | 6.12e-11 |
| GO:0034703 | 6.40e-11 |
| GO:0042995 | 7.17e-11 |
| GO:0097060 | 8.05e-11 |
| GO:0022835 | 2.26e-11 |
| GO:0022824 | 2.26e-11 |
| GO:0005230 | 2.75e-11 |
| GO:0022834 | 3.12e-11 |
| GO:0015276 | 3.12e-11 |
| GO:0019905 | 3.39e-11 |
| GO:0005244 | 3.88e-11 |
| GO:0022843 | 3.98e-11 |
| GO:0030594 | 5.26e-11 |
| GO:0030276 | 5.93e-11 |
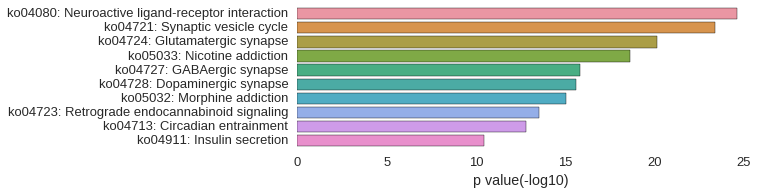
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG12488
TTTAATTCTTTAGAAAAGTGTTGATACATACATATGTGAAGAAACTGAACGATTGAATGAAAACGAAGAC
TAAAAAATGGATTTTTGTTGTTTAATTCAAAACAGCATGGGCGGCTCCTAACATATCACTTTCATTAATG
AAAACCAATGATTTGAGATGAAGCTTTCTCCAATGGGATGTCACTGCTGAAATGACATTGTGACTTTTTT
TTTCTCCCAAGCTGTACGTCACAAAGCTAAATATACTAATAAAACCGTG
TTTAATTCTTTAGAAAAGTGTTGATACATACATATGTGAAGAAACTGAACGATTGAATGAAAACGAAGAC
TAAAAAATGGATTTTTGTTGTTTAATTCAAAACAGCATGGGCGGCTCCTAACATATCACTTTCATTAATG
AAAACCAATGATTTGAGATGAAGCTTTCTCCAATGGGATGTCACTGCTGAAATGACATTGTGACTTTTTT
TTTCTCCCAAGCTGTACGTCACAAAGCTAAATATACTAATAAAACCGTG