ZFLNCG13198
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR023145 | heart | normal | 252.21 |
| ERR023146 | head kidney | normal | 85.75 |
| SRR592703 | pineal gland | normal | 54.10 |
| SRR592702 | pineal gland | normal | 50.91 |
| SRR1647682 | spleen | normal | 50.60 |
| SRR592701 | pineal gland | normal | 45.33 |
| ERR023143 | swim bladder | normal | 26.29 |
| SRR1647683 | spleen | SVCV treatment | 20.17 |
| SRR1647684 | spleen | SVCV treatment | 17.49 |
| SRR516122 | skin | male and 3.5 year | 16.96 |
Express in tissues
Correlated coding gene
Download
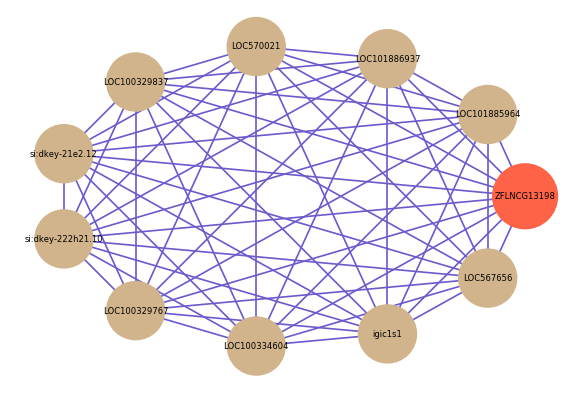
| gene | correlation coefficent |
|---|---|
| LOC101885964 | 0.74 |
| LOC101886937 | 0.72 |
| LOC570021 | 0.71 |
| LOC100329837 | 0.70 |
| si:dkey-21e2.12 | 0.70 |
| si:dkey-222h21.10 | 0.68 |
| LOC100329767 | 0.68 |
| LOC100334604 | 0.68 |
| igic1s1 | 0.68 |
| LOC567656 | 0.67 |
Gene Ontology
Download
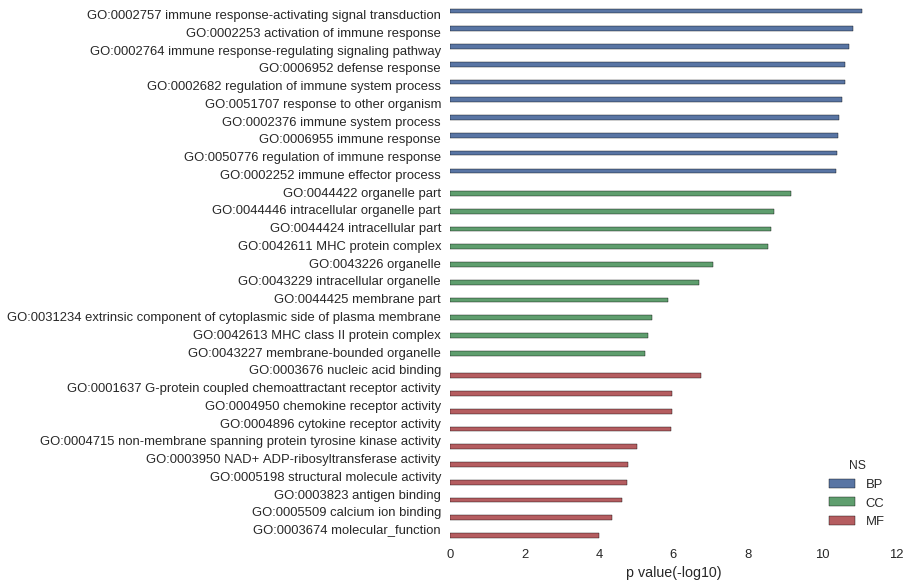
| GO | P value |
|---|---|
| GO:0002757 | 8.42e-12 |
| GO:0002253 | 1.50e-11 |
| GO:0002764 | 1.92e-11 |
| GO:0006952 | 2.41e-11 |
| GO:0002682 | 2.45e-11 |
| GO:0051707 | 2.85e-11 |
| GO:0002376 | 3.55e-11 |
| GO:0006955 | 3.76e-11 |
| GO:0050776 | 4.09e-11 |
| GO:0002252 | 4.13e-11 |
| GO:0044422 | 6.85e-10 |
| GO:0044446 | 1.94e-09 |
| GO:0044424 | 2.32e-09 |
| GO:0042611 | 2.82e-09 |
| GO:0043226 | 8.66e-08 |
| GO:0043229 | 1.99e-07 |
| GO:0044425 | 1.36e-06 |
| GO:0031234 | 3.82e-06 |
| GO:0042613 | 4.80e-06 |
| GO:0043227 | 5.72e-06 |
| GO:0003676 | 1.80e-07 |
| GO:0001637 | 1.12e-06 |
| GO:0004950 | 1.12e-06 |
| GO:0004896 | 1.16e-06 |
| GO:0004715 | 9.57e-06 |
| GO:0003950 | 1.71e-05 |
| GO:0005198 | 1.81e-05 |
| GO:0003823 | 2.41e-05 |
| GO:0005509 | 4.58e-05 |
| GO:0003674 | 9.91e-05 |
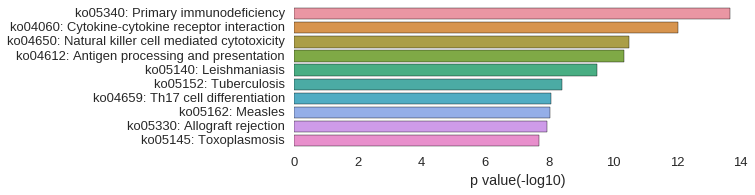
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG13198
ATTACACTGAAATCTGATGTACATAGCATCAAGCATAAACAAACACCAGACACATTTTTTACACAGCTTC
AGCCTTTGAGAAGTTACATTCACACACAATTACACAGAGTCAGTTATTTGTGAATATTTGCATCATGTTA
TTGATTGTACAGATTATGCTTGTATGGCACTAAACAGCACATAAACTGGCTAAATATATTCATTAACCAA
GGGTTGAAC
ATTACACTGAAATCTGATGTACATAGCATCAAGCATAAACAAACACCAGACACATTTTTTACACAGCTTC
AGCCTTTGAGAAGTTACATTCACACACAATTACACAGAGTCAGTTATTTGTGAATATTTGCATCATGTTA
TTGATTGTACAGATTATGCTTGTATGGCACTAAACAGCACATAAACTGGCTAAATATATTCATTAACCAA
GGGTTGAAC